All published articles of this journal are available on ScienceDirect.
Time Series Prediction of Viable Embryo and Automatic Grading in IVF using Deep Learning
Abstract
Background:
The process of In Vitro Fertilization (IVF) involves collecting multiple samples of mature eggs that are fertilized with sperms in the IVF laboratory. They are eventually graded, and the most viable embryo out of all the samples is selected for transfer in the mother’s womb for a healthy pregnancy. Currently, the process of grading and selecting the healthiest embryo is performed by visual morphology, and manual records are maintained by embryologists.
Objectives:
Maintaining manual records makes the process very tedious, time-consuming, and error-prone. The absence of a universal grading leads to high subjectivity and low success rate of pregnancy. To improve the chances of pregnancy, multiple embryos are transferred in the womb elevating the risk of multiple pregnancies. In this paper, we propose a deep learning-based method to perform the automatic grading of the embryos using time series prediction with Long Short Term Memory (LSTM) and Convolutional Neural Network (CNN).
Methods:
CNN extracts the features of the images of embryos, and a sequence of such features is fed to LSTM for time series prediction, which gives the final grade.
Results:
Our model gave an ideal accuracy of 100% on training and validation. A comparison of obtained results is made with those obtained from a GRU model as well as other pre-trained models.
Conclusion:
The automated process is robust and eliminates subjectivity. The days-long hard work can now be replaced with our model, which gives the grading within 8 seconds with a GPU.
1. INTRODUCTION AND BACKGROUND
Infertility has become a global health issue, and global statistics indicate that 8-10% of couples suffer from infertility [1]. According to the World Health Organization (WHO), 1 out of every 4 couples is affected by infertility [2]. As a result, there has been a drastic increase in the number of couples seeking In Vitro Fertilization (IVF) worldwide. Globally, approximately 5 million children have been born with the help of IVF [3]. However, some couples remain childless even after several IVF cycles. Due to the uncertainty in the viability of the embryos, multiple embryos are transferred for maximizing the odds of pregnancy, and hence women undergoing IVF face greater risks of multiple pregnancies along with financial burden.
The typical IVF cycle starts with collecting multiple samples of eggs and sperms from the parents, and the fertilization takes place in a controlled environment. The fertilized egg, now called an embryo, is kept inside an incubator for 5 to 6 days where its development stages are meticulously examined by the embryologists. They assign grades to the embryo as per its feature and efficient development with time. The grades are assigned on all days of the cycle at fixed hours measured with the unit of HPI, which stands for hours post-insemination. Ultimately, the sample with the best grading is considered viable for embryo transfer [4]. Incubators with built-in time-lapse monitoring can enable non-invasive embryo assessment. Comparatively, fine-grained detail, inspiring significant interest in applying embryo―morphokinetics is needed to score and prioritize embryos. Morphokinetics comprise the timing and morphologic appearance of embryos as they grow and pass through a series of sequential developmental stages. Currently, embryologists must perform the morphokinetic analysis manually. Better accuracy of automated grading is expected as compared to the human embryologist.
Visual morphology assessment is routinely used for evaluating embryo quality and selecting human blastocysts for transfer after fertilization and complete development of the embryo. However, it is quite often that there is a difference of opinion between embryologists since this process is based on observation of the changes occurring in the embryo samples [5, 6]. As a result, the success rate of IVF remains low, and patients often have to undergo more than one cycle of treatment before a healthy pregnancy is assured. The IVF procedure can cause emotional and psychological stress to the patients. To overcome such uncertainties in embryo quality, a method is adopted which involves transferring more than one embryo to the uterus. This increases the risk of multiple births. Such a pregnancy with multiple fetuses may lead to complications such as early labor and low birth weight than a pregnancy with a single fetus does. Moreover, multiple pregnancies also carry health risks to the mother and child. Also, the grading system consists of maintaining long manual records of analyzing the multiple samples at different times of the development cycle. This process is very tedious and time-consuming and not fit for the busy environment of the IVF labs. To combat these issues, it becomes necessary to bring about changes in the current techniques, thus making this procedure more efficient.
The Artificial Intelligence-driven approach provides an efficient way to assess embryo quality and reveals new, potentially personalized techniques to select embryos. By introducing deep learning into the field of IVF, we can automate and standardize the grading process of embryos, a process that is very dependent on subjective human judgments. Tsung-Jui Chen et. al. used a deep learning-based approach to be applied to a large dataset of embryo images [7]. A CNN model was implemented using a pre-trained ResNet50 architecture for predicting the grades. The outcomes of the study involved an average predictive accuracy of 75.36% and exhibited the success of employing deep learning for embryo grading.
Inspired by such studies, the main aim of this research paper is to explore the capabilities of deep learning and artificial intelligence and implement a method based on deep learning to automatically grade the structural appearance of human blastocysts using LSTM-CNN and thus, generate results that are above human level accuracies to identify healthy embryos for IVF.
Some of the main contributions of the paper are as follows:
- Understanding the parameters necessary for embryo grading.
- Proving the importance and impact of time series prediction for grading the embryo.
- Implementing an LSTM-CNN Network on the dataset.
- Analysis of outcomes of different CNN pre-trained models on training and validation accuracy.
- Implementing GRU, which is also a well-known time series predictor like LSTM, and analyze its results
- Comparing the performance of embryologist grading and grading of our proposed model in terms of accuracy and time taken for the entire grading process
The present study is organized as follows: In Section 2, we have presented the grading method used by the fertility center from where the dataset was collected. Here, we have also presented related works using deep learning for embryo grading. A brief explanation of the embryo development through different stages, which is essential to understand the automation in the work done is given. In Section 3, we have described our proposed methodology for embryo grading using deep learning. In Section 4, we have described the dataset used, followed by a detailed discussion on the experimental results obtained in Section 5. Finally, in Section 6, we have presented the conclusions offered by the work done.
2. LITERATURE REVIEW
This section deals with the detailed discussion of the embryo development stages which are essential for an intuitive approach towards automating the process. The development stages are distributed in 5-6 days and are analyzed on all the days for predicting the final grade. In the later subsections, we have reviewed the various proposed approaches adopted by researchers with a similar aim.
2.1. Stages of Embryo Development
After the egg sample is fertilized with the collected sperm sample of the patient, they are kept under supervision inside the incubator where the entire development process from the single fused cell till the blastocyst stage takes place. The development stages of the embryo in the incubators are as follows [8]:
- (Day 0 – 1) Pronuclear Stage
During the pronuclear stage, the sperm and the egg unite to form one single cell. The nucleus of each of these two gametes combines to form pronuclei. This is essential for an embryologist to deduce that the fertilization has successfully taken place. Fig. (1a) shows the image of day 1 of the development process, just after fertilization.
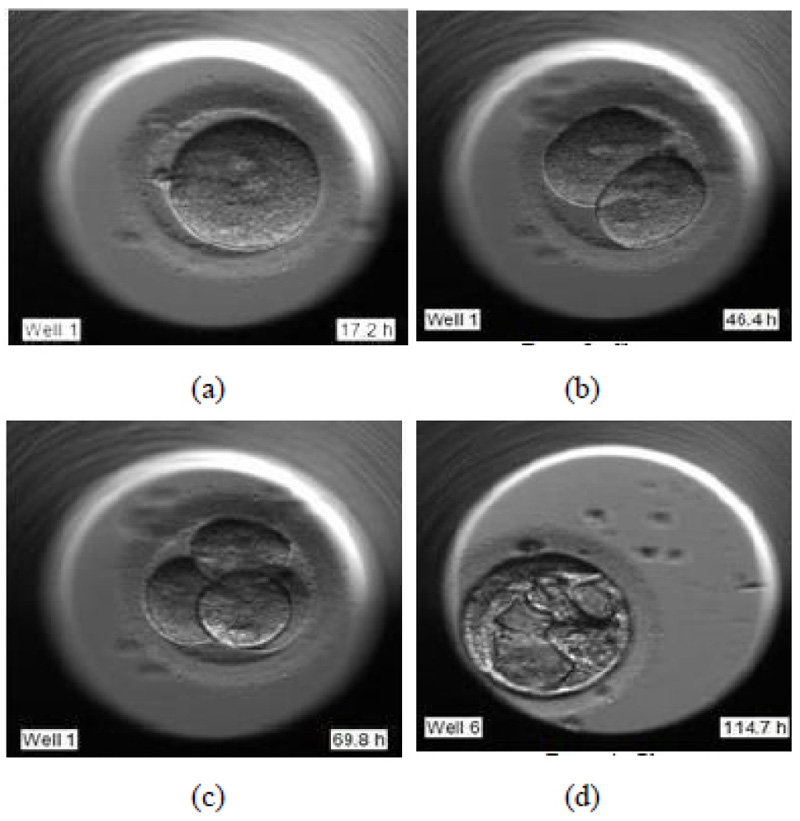
- (Day 1 – 3) Cleavage stage
During the cleavage stage, the cell divides itself into multiples of two. These two cells further split to form four and so on. They are called the cleavage stage embryos. In Fig. (1b) the cell division is visible clearly, where the two cells are formed.
- (Day 3 – 5) Morula stage
Fig. (1c) shows the further splitting of two cells into four. Usually, after day 3, the cells which have so far splitted themselves into multiple cells undergo compaction which is a part of the morula stage. This results in a solid, compact mass of multiple cells.
- (Day 5 – 6) Blastocyst stage
The blastocyst stage is one of the most important stages. The cells undergo expansion and a visible transformation into its 2 expected components: Inner Cell Mass (ICM) and the Trophectoderm, which further develops to become the fetus and placenta respectively. Fig. (1d) shows the formation of ICM and trophectoderm during the blastocyst stage.
2.2. Embryo Grading
Morphology is the primary method adopted by embryologists for the assessment of the development of the embryo. During IVF, the embryos are cultured till the day of transfer and their quality is tested during each day of the grading cycle. It is essential to evaluate the maturity of the eggs before fertilization. To assign a qualitative measure to the embryos, various grading methods have been introduced over the years. However, an ideal universal grading system has not been established, indicating the complexity and the dynamic nature of the embryo along its development stages. As a result, many fertility centers adopt grading schemes based on their embryo analysis methods.
The dataset used for our study was procured from Nova IVF Fertility Center in Ahmedabad, Gujarat, India [9]. The embryologists of this fertility center have adopted a typical method of evaluating the embryos. The embryologists observe the development stages of the embryo by analyzing the features as the embryo develops from day 1 till day 5-6. The embryologist assigns grades to the embryo from the very beginning when the samples are collected. The sperm and the egg samples are also individually graded before fertilization.
Once the fertilization is done, the development stages are constantly analyzed at fixed HPIs starting from day 1 till day 5 or 6 or even before, based on the health and viability of the embryo. They maintain a handwritten record of the grades in a tabular form. A final grade is assigned on the last day before taking it out of the incubator. Based on the final grades, the viability of the embryo is predicted, and the best sample or more are selected and transferred into the mother‘s womb. The grading of the embryologists was made available from the hospital. The results were handwritten in the form of manual grades assigned to all patients and all their samples. The grading scheme adopted [10] is, as shown in Table 1.
| Grade | Embryo Quality |
|---|---|
| A | Best |
| B | Good |
| C | Fair |
| D | Poor |
| E | Non-Viable |
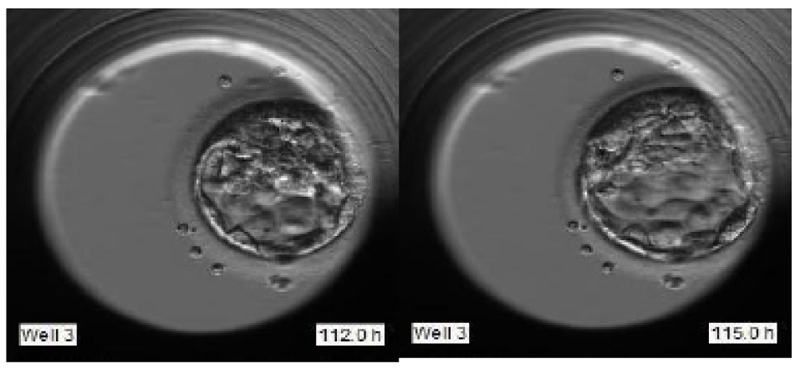
These grades are allotted based on various features like Degree of expansion, Inner cell mass (ICM), and Trophectoderm (TE), as portrayed in Fig. (2).
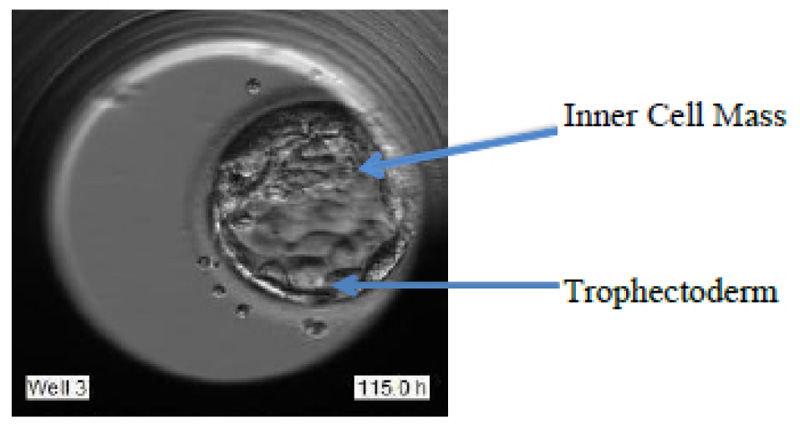
The best grade embryo has the following features:
- The large expansion of blastocyst as indicated in Fig. (3).
- A large number of uniformly-sized cells in the trophectoderm.
- Fat looking ICM with many cells tightly packed together.
2.3. Related Works
The grading process and the evaluation of the most viable embryo for embryo transfer are highly subjective along with the complexity that arises due to the dynamic nature of embryo development. Several approaches have been proposed to automate the grading process of the embryos and the selection of the healthiest one for embryo transfer. With the advent of machine learning applications in various domains, the field of embryology has had its impact too.
One of the known methods of automating the grading process by introducing deep learning was with the use of Convolutional Neural Networks, which are extremely powerful neural networks, especially for dealing with images. The work cited in the paper by Chen et al. [7]. has implemented a deep learning technique on a large dataset of microscopic images. A Convolutional neural network has been applied to the dataset, and a ResNet50 architecture has been used to tune the parameters. The labeling of the embryo images has been done using Gardner‘s grading system, which is one of the many well-known methods for grading embryos. The blastocyst development, ICM quality, and TE quality were the focal points of analysis and the results indicated an accuracy of 75.36% for the automated grading of the dataset.
A similar CNN based methodology was proposed in the paper [11] for embryo selection and classification. As per the previous discussion, Gardner‘s grading standards [12] were adopted here too for the labels of the dataset. Accuracy, sensitivity, and specificity are the 3 evaluation criteria considered and the results obtained by the method are 78.14%, 87.0%, and 68.82% respectively, for the three criteria. Besides the low results, the research consists of some other limitations too. The image analysis done is only favorable for static 2D images. They have concluded that developing a model with considerations of the temporal development of the embryo using a time-lapse video dataset can provide much better results.
A different kind of approach was adopted in the paper [13]. The developed method was named STORK and it makes use of Deep Neural Networks to perform automatic embryo grading for the selection of the highest quality of embryos. Veeck and Zaninovic grading method [14] was followed. The novel feature of STORK is that instead of focusing on the predetermined features only, it analyzes the entire image of the embryo. Hence CNN is given the power to analyze all those features which were not being assessed or not considered important earlier. The dataset consists of 50,000 time-lapse images of human embryos. The STORK framework, based on Google’s Inception model has been used for blastocyst quality prediction. A collection of 2182 embryos have been used to generate a decision tree using a CHAID algorithm that relates the embryo quality with the patient’s age. The accuracy obtained was 97.53%. Another Region-based CNN based approach proposed in [15] took into account the position change of the cells with the help of the Kalman Filter. To correlate the changes in cells in the video frames a Hungarian algorithm was used. The proposed model can track up to 4 cells. Adding to the list, another study done in the paper [16] makes use of cloud computing. Microsoft Azure APIs are used for the classification of the IVF images. The proposed model achieved a recall rate of 89.2% and a precision of 85.7%.
Looking at the recent advances in the research in this field, it is evident that interpretation of the quality of the embryos is very complicated, and giving any assignments based on visual morphology without considering the time stamp or the analysis of the temporal information is just a conjecture. The theory has been proved by analyzing the embryos at different times of the same day and obtains non-obvious results due to their dynamic nature. Analyzing the embryos multiple times on the same day again brings in other issues like the sudden shoot in time spent by the embryologists. There is also an increased risk added due to the removal of embryos from the incubator for analysis in the absence of time-lapse microscopy. Time-lapse microscopy was introduced to serve the purpose of enabling the embryologists to leverage the temporal information with eliminated risks. But the time consumption in analyzing multiple samples of this 5-6 day process of thousands of patients is quite impractical.
The latest algorithms have effectively utilized the data from time-lapse microscopy for automating the process. The algorithms focus on the multiple events in the development cycle of the embryo, along with their timestamps. A similar approach is discussed in the paper [17]. They have made use of the time-lapse imaging for grading the morphological appearance of human blastocysts with the implementation of CNN as well as RNN. CNN predicts inner cell mass (ICM) and trophectoderm (TE) grades from a single image frame, and then RNN takes into account the temporal information from multiple image frames. The accuracy result for ICM grading is 65.2% and 69.6% for TE. There was an improvement in accuracy as ICM 7.2%; TE 5.1% as compared to the results obtained from the CNN model on static images.
In this paper, we have proposed a methodology that provides a fully automated time-series grading of the embryos along with higher accuracy for the grading and detection of the most viable embryo for transfer.
3. DATASET DETAILS AND PREPROCESSING
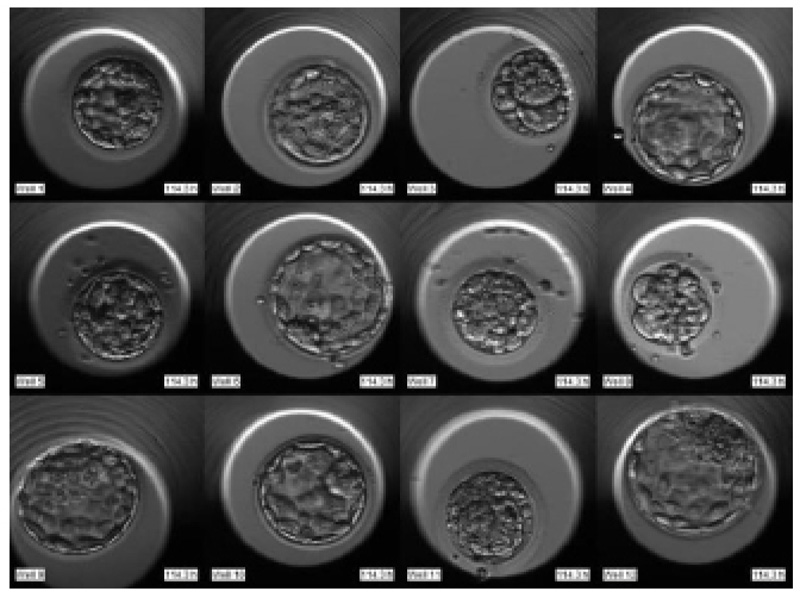
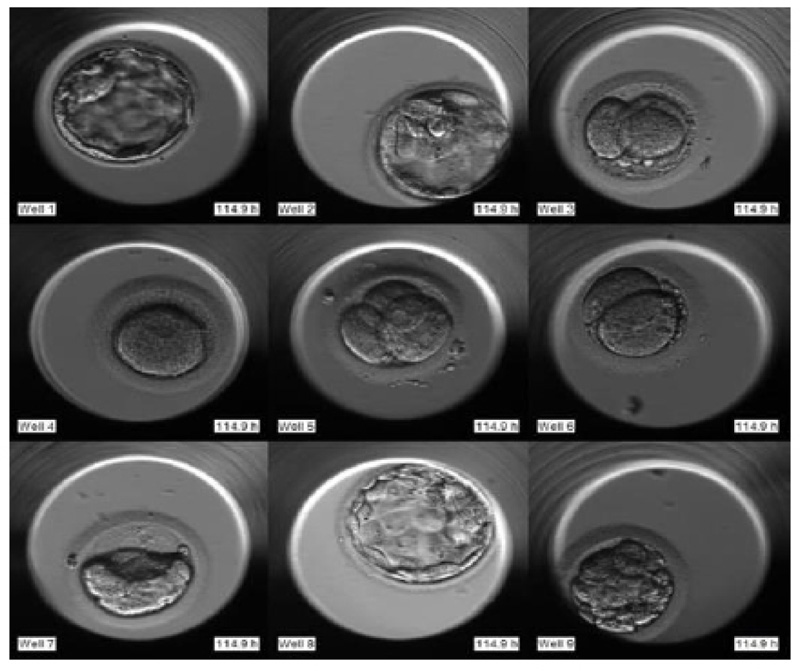
The required dataset was obtained from Nova IVF Fertility, Ahmedabad [9]. The hospital provided the video dataset of multiple patients with multiple samples from the embryoscope. We received data of 60 patients with 10-30 samples each. Hence a total of 803 labeled samples were obtained from the hospital. Since each patient had multiple samples, the video dataset of individual patients was in the form of a grid of 3x4 if the patients had more than 9 samples and 3x3 otherwise. Fig. (4) shows the image frame extracted from the video dataset of a patient with 12 samples arranged in a 3x4 grid, and Fig. (5) shows the dataset of a patient with 9 samples arranged in 3x3 grids. Each patient had multiple samples, so each sample of each patient was extracted separately as well.
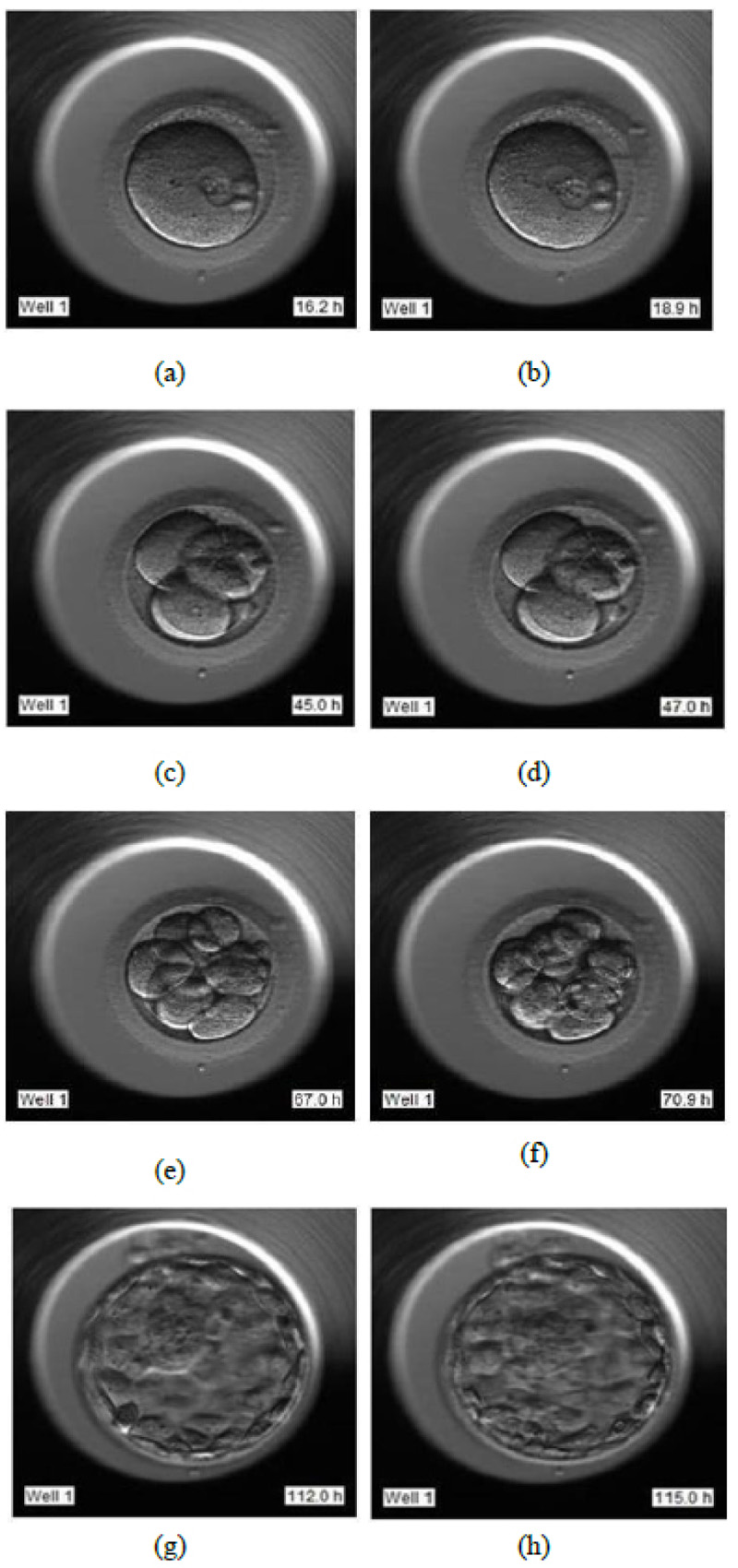
The video footage captures the entire development process from day 1 till day 5 or 6. The video of this days-long process was compressed to 2-3 minutes, and frames were captured at fixed intervals in terms of HPI.
- Day 1: Image frames extracted in the interval of 16 HPI to 19 HPI as shown in Fig. (6a and 6b), respectively
- Day 2: Image frames extracted in the interval of 44 HPI to 47 HPI as shown in Fig. (6c and 6d) respectively
- Day 3: Image frames extracted in the interval of 67 HPI to 71 HPI as shown in Fig. (6e and 6f) respectively
- Day 5: Image frames extracted in the interval of 112 HPI to 115 HPI as shown in Fig. (6g and 6h) respectively.
These frames correspond to images from day 1 to day 5 of individual samples of individual patients. 5458 image frames were extracted in total, which comprised all the image frames of days 1, 2, 3, and 5 of all 803 samples. The extracted images were of the size of 250x250 pixels. The image extraction is essential as the first step of the data processing which will serve as an input to our LSTM- CNN model. The images were further cropped to 197x200 pixels for extracting the area of interest.
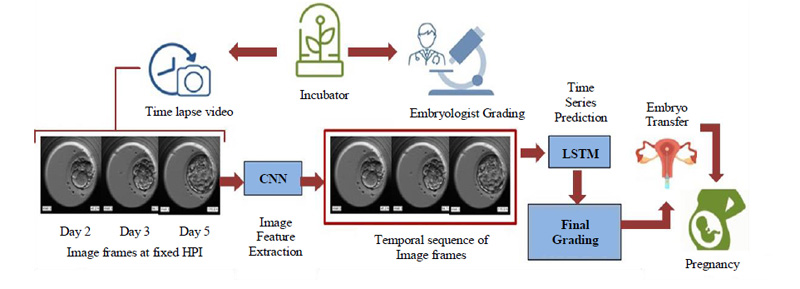
4. PROPOSED METHOD
The dynamic nature of the embryos over even short periods highlights the importance of temporal information and makes the use of time series prediction inevitable. By linking time stamps to the dynamicity of the embryo, we can keep track of the process and evaluate the sequential growth accordingly. Considering these important factors our model rightly makes use of time series prediction with the help of Long Short Term Memory (LSTM), which is a special kind of Recurrent Neural Network (RNN) favored for dealing with sequential data. The basic flow of the proposed and developed model is as shown in Fig. (7).
The embryoscopic time-lapse videos of the development process are recorded and image frames at fixed HPI are obtained which can be fed to Convolutional Neural Network (CNN) for image feature extraction. These image features in sequential order are fed to Long Short Term Memory (LSTM) and based on the time stamps, the final grading is performed. Based on the final grades, the most viable embryo is selected for embryo transfer in the prospective mother’s womb leading to a successful pregnancy. The proposed methodology will be discussed in detail in further subsections.
4.1. Image Feature extraction using CNN
Convolutional Neural Network (CNN) is proficiently known for its high accuracy in image recognition and classification. As a part of computer vision, Transfer learning [18] is one of the very efficient and time-saving methods while developing our model. Through Transfer Learning, you can directly make use of already developed models that are trained with high accuracies on huge datasets. These models are known as Pre-Trained models. They can be readily used without the need to develop the entire CNN architecture. They provide high accuracy along with saving a lot of time. We have chosen various available pre-trained models and used them along with LSTM in our model.
The cropped image frames of size (197x200x3) serve as the input to the Pre-trained CNN model, which extracts features from the image frames. The output shape of the model is (6x6x512). 512 is the number of features extracted from the images.
4.2. Time Series prediction using LSTM
As mentioned in Section 1, time series prediction plays a significant role in determining the grades of the embryos. LSTM plays an important role in extracting input patterns when the input extends over long sequences. Since the gated architecture of LSTM is capable of manipulating memory states, they are suitable for such problems. It remembers every bit of information through time and is hence, useful in time series prediction due to its ability to remember previous inputs [19].
After having extracted the features using the Pre-trained CNN model, the image features arranged in a sequence are further fed to the LSTM layer for leveraging the temporal information. The features are arranged in a sequence of images from day 2, day 3 till day 5. The output of the pre-trained model must be passed through a flatten layer, as well as a, reshape layer before serving as an input to the LSTM layer. The LSTM input shape is 3 dimensional, with the dimensions being the batch size, time steps, and the input units. In our case, the time steps are 3 correspondings to day 2, day 3, and day 5 images in the sequence and the input units are the number of features obtained from the pre-trained model. The input of shape (None, 3, 18432) obtained after the flatten and reshape layers is fed to the LSTM layer. The output of the LSTM layers is of the shape (None, 128), which is then passed through 2 dense layers. The final layers with the output shape of 5 imply the 5 possible outcomes of the grading ranging from 0 to 4. The final output is the predicted grade based on the sequence of embryo growth fed to the LSTM layer. LSTM processes this temporal information and predicts the grading of the embryo.
4.3. Loss Function
Cross entropy is the loss function used for multi-class classification problems and is based on the concept of maximum likelihood. In cross-entropy, the difference between the predicted and actual probabilities is averaged and the probability distribution is summarized to minimize the score to the ideal value of cross entropy which is 0.
The loss functions ‘Categorical cross-entropy’ as well as ‘Sparse categorical cross entropy’ is commonly used. Categorical cross-entropy requires one-hot encoding of the labels. For our model, we have chosen ‘Sparse categorical cross entropy’ loss function [20] to rule out the requirement of one hot encoding of our labels.
5. RESULTS
The LSTM-CNN model was trained on a GPU as per the following experimental setup details:
- Train-Validation split: 80% - 20%
- Batch Size: 8
- Optimizer: Adam
- Learning rate: 0.0001
- Loss Function: Sparse categorical Cross entropy
- RAM requirements: 25 Gb
Various Pretrained CNN models were implemented on the embryo image dataset, and their performance was compared. The parameters batch size and the ratio of training to validation data were made to vary for the performance analysis. Of these models, the best one was selected and implemented on the dataset along with LSTM, and very promising results were obtained, proving the capability of LSTM in time series prediction.
5.1. Model Specifications
The model was trained using numerous pre-trained models like VGG19 [21], Xception [22], MobileNet [23], DenseNet121 [24] and Inception [25]. We also trained the model using GRU, which is also used for time series prediction like LSTM [26]. As the name suggests, these models are already trained and they provide a very high accuracy when used with a large amount of image data. Since all these pre-trained models are favorable while dealing with images, almost all the models have given 100 percent accuracy.
Table 2 indicates that the model was also trained by varying the various parameters like batch size and train – validation split ratio. The batch size of 8 was found to give the best performance and the best accuracy was provided by VGG16 [27].
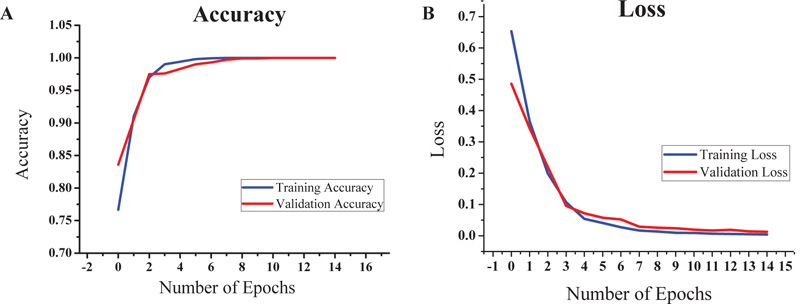
Based on the promising results given by VGG16, we decided to focus on it for further study. The VGG16-LSTM model and an accuracy of 100% was obtained for both training and validation dataset. The train-validation split was 80-20 in this case and the steps per epoch were also set accordingly. The batch size was kept as 8. The graphs of accuracy and loss obtained after running the VGG16-LSTM model are shown in Figs. (8a and 8b), respectively.
Like LSTM, GRU (Gated Recurrent Unit) [26] is also a well-known model for time series prediction. In contrast to LSTM, which has three gates namely the input gate, forget gate, and the output gate, GRU has two gates, which are the reset gate and the update gate. LSTM is preferred while dealing with large data and when there is a requirement of high accuracy. On the other hand, GRU is generally preferred when the data is less and accuracy is of less importance.
The model achieved satisfactory accuracy within 15 epochs. Fig. (9) indicates the distribution for different batch sizes and fixed train to the validation data ratio of 70-30. Fig. (10) ultimately shows the Training and Validation accuracy of different CNN pre-trained models with LSTM. Table 2 presents the accuracy of the models under different values of batch size and train to validation data ratio.
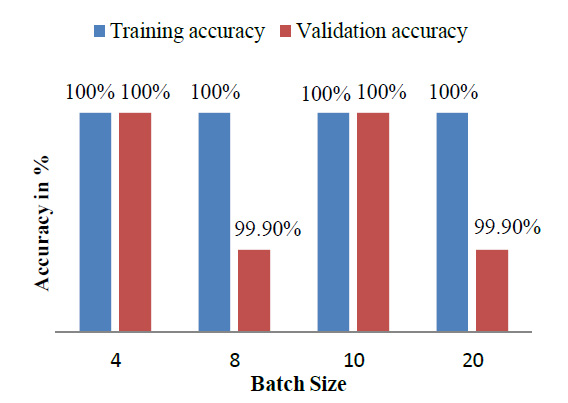
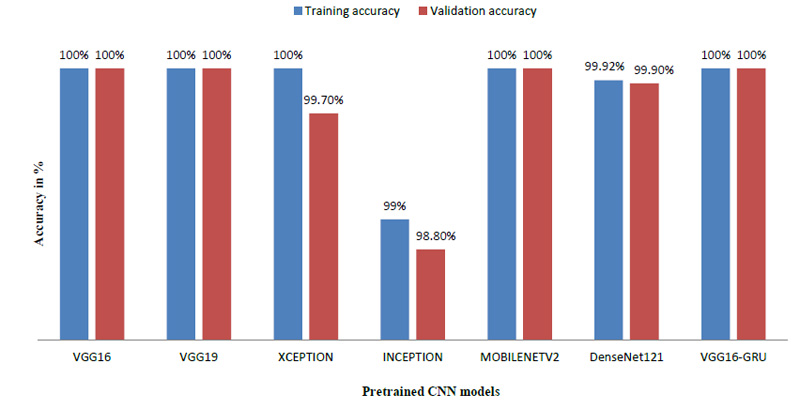
| Model | Train-Validation split (%) | Batch Size | Training Accuracy (%) | Validation Accuracy (%) |
|---|---|---|---|---|
| VGG16-LSTM | 80-20 | 10 | 100 | 100 |
| VGG16-LSTM | 80-20 | 8 | 100 | 100 |
| VGG19-LSTM | 80-20 | 10 | 100 | 100 |
| Xception-LSTM | 80-20 | 10 | 100 | 99.7 |
| Inception-LSTM | 70-30 | 10 | 99 | 98.8 |
| MobileNetV2-LSTM | 70-30 | 10 | 100 | 100 |
| DenseNet121-LSTM | 80-20 | 8 | 99.2 | 99.9 |
| VGG16-GRU | 80-20 | 8 | 100 | 100 |
| Sample | Patient 1 | Patient 2 | Patient 3 | |||
|---|---|---|---|---|---|---|
| LSTM Model Grades | Manual Srades | LSTM Model Grades | Manual Grades | LSTM Model Grades | Manual Grades | |
| 1 | 4 | 4 | 4 | 4 | 4 | 4 |
| 2 | 4 | 4 | 2 | 2 | 2 | 4 |
| 3 | 4 | 4 | 4 | 4 | 2 | 2 |
| 4 | 4 | 4 | 4 | 4 | 4 | 1 |
| 5 | 4 | 4 | 4 | 4 | 4 | 4 |
| 6 | 4 | 2 | 4 | 4 | 4 | 4 |
| 7 | 3 | 3 | 4 | 4 | 4 | 4 |
| 8 | 4 | 4 | 2 | 2 | 4 | 4 |
| 9 | 3 | 4 | 4 | 4 | 4 | 4 |
| 10 | 4 | 4 | 4 | 4 | 4 | 4 |
5.2. Test Results
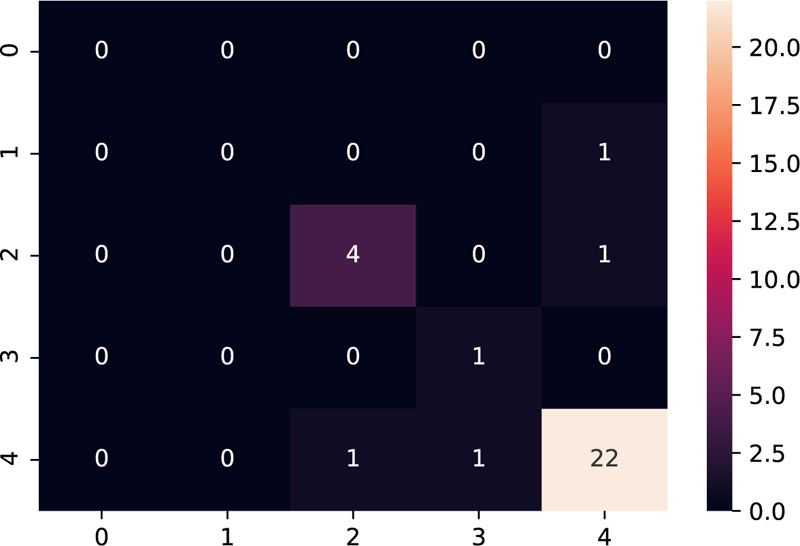
The model was tested on the dataset of three patients having 10 samples each. Table 3 shows the results after the model was implemented on the test data. The grades predicted by the model have been compared with the grades given manually by the embryologists. The model predicts grades of 26 samples correctly out of 30 samples, thus providing accuracy close to the human level.
Amongst the multiple samples collected from the patients for selecting the best embryo, a majority of the samples turned out to be non-viable, as expected, which is indicated as grade 4 as per our grading scheme. The confusion matrix [28] as shown in Fig. (11), supports this fact by indicating the maximum diagonal entry for grade 4 and the model has predicted it accurately too. The non-diagonal entries indicate that there have been 4 instances where a grading mismatch is encountered. However, this is a result of certain anomalies in the video dataset, which needs to be taken care of while dealing with the dataset to make the model more robust.
The video should be recorded until the instance when it has to be taken out of the incubator. If the video could not capture the final blastocyst then one sort of anomaly can occur. Various such anomalies can be avoided if we ensure that our time-lapse video dataset is complete as a whole which is the basic and the essential requirement.
5.3. Loss Function
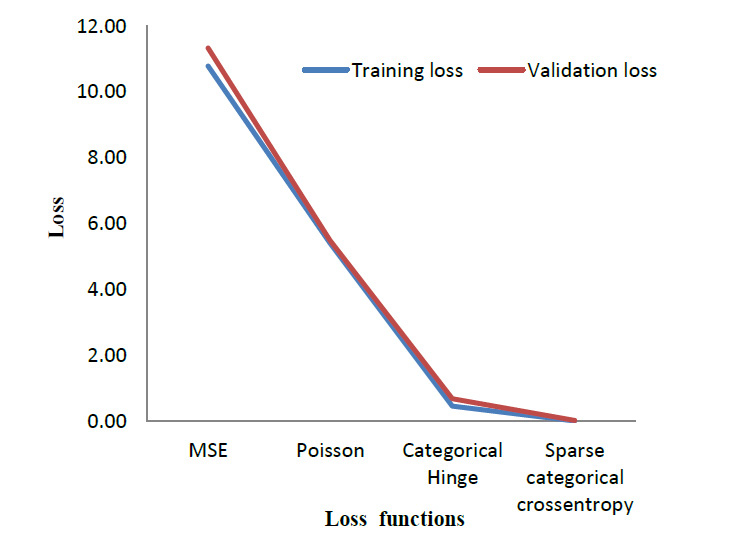
To evaluate the performance of the cross-entropy loss function, its performance was compared with three other loss functions. Table 4 lists out the results obtained when the model is trained using different loss functions. Mean Square Error (MSE) [29] and Poisson [30] are the most commonly used loss function for regression models. Whereas Categorical Hinge [31] and Sparse Categorical Cross entropy [32] are loss functions used for multi-class classification problems. The suggested cross-entropy loss function provides the best results. As mentioned in Section 3.2, Sparse Categorical cross entropy does not require one-hot encoding of labels [33]. Hence, it also performs better in terms of memory usage since it makes use of a single integer for a class, rather than the whole vector. Fig. (12) gives a visual overview of the results so obtained.
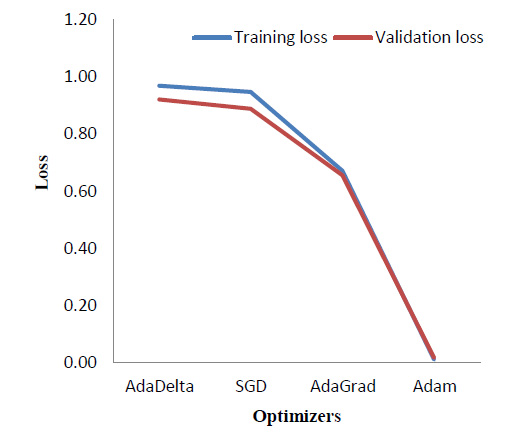
5.4. Optimizers
Various optimizers were implemented and a comparison of the results in terms of the training and validation loss was obtained as shown in Table 5. Fig. (13) correspondingly indicates the comparison graphically.
As seen in Fig. (13), Adam [34] optimizer performs the best, providing a significantly reduced loss as compared to the other three optimizers, which are - AdaDelta [35], Stochastic Gradient Descent (SGD) [36], and AdaGrad [37]. As theoretical support, Adam is the most widely used optimizer. It combines the benefits of Stochastic Gradient Descent and AdaGrad.
5.5. Timing
The proposed method was tested using a CPU as well as a GPU as the hardware accelerator. Table 6 lists the total execution time taken by the proposed CNN - LSTM model to predict the grades. GPU performs the entire process in just 8 seconds which is very remarkable as compared to the days-long manual morphokinetics performed till date for the same.
| Loss Function | Training Accuracy | Validation Accuracy | Training Loss | Validation Loss |
|---|---|---|---|---|
| MSE | 20.83% | 46.40% | 10.78 | 11.32 |
| Poisson | 23.30% | 32.90% | 5.40 | 5.48 |
| Categorical Hinge | 40.50% | 65.80% | 0.46 | 0.68 |
| Sparse Categorical Cross entropy | 100.00% | 100.00% | 0.01 | 0.02 |
| Optimizer | Training Accuracy | Validation Accuracy | Training Loss | Validation Loss |
|---|---|---|---|---|
| AdaDelta | 67.50% | 70.30% | 0.97 | 0.92 |
| SGD | 67.10% | 70.40% | 0.95 | 0.89 |
| AdaGrad | 74.48% | 76.40% | 0.67 | 0.66 |
| Adam | 100.00% | 100.00% | 0.01 | 0.02 |
| Platform | Total Time (sec) |
|---|---|
| CPU | 240 |
| GPU | 8 |
6. DISCUSSION
The above results suggest that the proposed model can predict embryo grades with acceptable accuracy. The model can provide steady results, and thus avoiding observer variances. The model is fully automated and does not require any manual processing of embryo images before prediction. As indicated in Table 2, VGG-16 [27] provides one of the best training and validation accuracies when used along with the LSTM layers. Moreover, a train to test split of 80 to 20, keeping a batch size of 8, was found to be most efficient in obtaining accurate results. Our model makes use of the loss function Sparse Categorical cross entropy [20]. Table 4 indicates that this loss function provides a 100% training and validation accuracy with negligible loss. As per Table 5, Adam optimizer [34] proves to be the best, providing 100% accuracy.
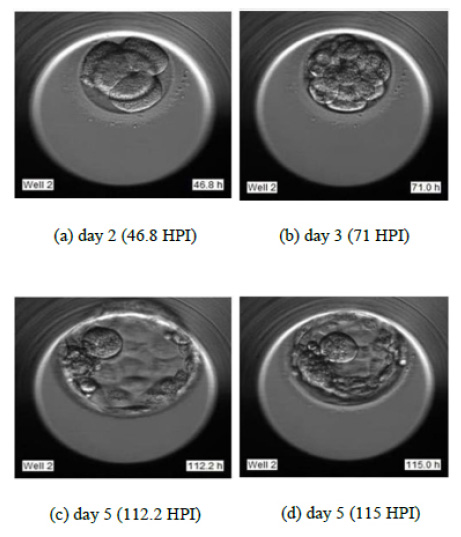
To justify the accurate grading, we have selected the sample images of the second sample of patient 2 as indicated in Fig. (14). Fig. (14) indicate the embryo samples observed at 46.8, 71, 112.2, and 115 HPI respectively. The embryo development stages on days corresponding to these HPIs appear very promising. This particular embryo sample has been considered viable by the embryologist and grade 2 has been assigned to this sample. Our LSTM-CNN model has given similar results.
The 4 misclassified samples are marked in bold in Table 3. While analyzing the discrepancy in the misclassified samples, the 6th sample of patient 1 has been assigned grade 2 by the embryologist. However, on verifying, it was found that for sample 6, the blastocyst stage has not been captured in the time-lapse video. Hence, due to the absence of day 5 images, a discrepancy occurred and grade 4 is assigned. In the case of patient 3, a visual analysis of samples 2 and 3 indicate that they are undergoing the same stages of development until the blastocyst stage. So their grades should be the same, which the model is predicting correctly. For the same patient, sample 4 does not reach the blastocyst stage until the end, and so the model predicts grade 4. Hence we have justification from the video data set for all the samples where there is a grading mismatch, as discussed in detail in the previous section.
CONCLUSION
The project has developed a fully automated grading system for embryos in IVF along with achieving high accuracy of 100 percent using the time-lapse images of the development of the embryo in the incubators. The method uses neural networks for analyzing the time series information along with the information of the image features and predicts the grades for the embryo after blastocyst. Morphokinetics for Embryo grading can be efficiently automated and the prediction of viable embryos can be made simpler, faster, and more accurate as compared to the manually performed morphokinetics at present. The comparison of various other pre-trained CNN models was also made from which VGG16, VGG19, Xception, and MobileNET gave promising results and 100 percent accuracy as well. A VGG16-GRU model was also implemented, resulting in 100 percent accuracy. For a complete analysis, various optimizers and loss functions were compared. Adam optimizer and Sparse categorical cross-entropy loss function were proved to be ideal for our model. The total time taken for the final grading of the embryo using a CPU and GPU were compared. With the help of a GPU, the days long grading process could be completed within just 8 seconds.
Results have proved that the usage of time-lapse images for analyzing the temporal information gives much better results as compared to single image evaluation. Time series analysis using LSTM has proved to give very promising results. The proposed CNN-LSTM model is fit to be deployed in the busy environment of the IVF labs and automate the highly subjective manual process. Further, after the GUI development, hospitals can easily use this model for a faster and more accurate grading system.
LIST OF ABBREVIATIONS
| CNN | = Convolutional Neural Network |
| GPU | = Graphics Processing Unit |
| GRU | = Gated Recurrent Unit |
| HPI | = Hours Post Insemination |
| ICM | = Inner Cell Mass |
| IVF | = In Vitro Fertilization |
| LSTM | = Long Short Term Memory |
| VGG | = Visual Geometry Group |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
Not applicable.
CONSENT FOR PUBLICATION
Informed consent was obtained from all participants.
AVAILABILITY OF DATA AND MATERIALS
The dataset was procured from Nova IVF Fertility, Ahmedabad, Gujarat, India. For the protection of doctor-patient confidentiality, the dataset cannot be shared.
FUNDING
None.
CONFLICT OF INTEREST
The author declares no conflict of interest, financial, or otherwise.
ACKNOWLEDGEMENTS
We would like to express our sincere gratitude towards Nova IVF fertility for helping us with the most crucial requirement of the research, which is the dataset.


