All published articles of this journal are available on ScienceDirect.
Identifying Skeletal Maturity from X-rays using Deep Neural Networks
Abstract
Skeletal maturity estimation is routinely evaluated by pediatrics and radiologists to assess growth and hormonal disorders. Methods integrated with regression techniques are incompatible with low-resolution digital samples and generate bias, when the evaluation protocols are implemented for feature assessment on coarse X-Ray hand images. This paper proposes a comparative analysis between two deep neural network architectures, with the base models such as Inception-ResNet-V2 and Xception-pre-trained networks. Based on 12,611 hand X-Ray images of RSNA Bone Age database, Inception-ResNet-V2 and Xception models have achieved R-Squared value of 0.935 and 0.942 respectively. Further, in the same order, the MAE accomplished by the two models are 12.583 and 13.299 respectively, when subjected to very few training instances with negligible chances of overfitting.
1. INTRODUCTION
The determination of bone age provides information about an individual’s structural and biological maturity. It can be used as a tool for clinical diagnosis of diseases associated with abnormally short or tall stature in children [1] or for forensic purposes. It can also prove to be useful in ascertaining the chronological age if accurate birth records are unavailable. Many deep learning applications have been successful in substituting the former methods.
Traditionally, the Tanner Whitehouse [2] and the Geurich and Pyle [3] methods are widely practiced in clinical assessment and diagnostics; however, these are labor-intensive and time consuming, vulnerable to observer’s mishandling. Predictive analysis is carried out on four major ossification regions in hand, namely epiphyses bone, medial carpal, radius, and the ulna. The first three regions drastically vary according to age, sex and ethnicity [4, 5]. The phalangeal analysis is the most suitable in children (above age 6 in females and above age 8 in males) and therefore, computer-aided medical diagnostic (CAD) systems [6-8] method can be deemed the best if applied. The associated techniques can pick out relevant aspects from the phalangeal region using a digital hand atlas. The same cannot be applied to children below the ages of 5-7 years since the presence of soft tissue makes the process of segmentation between epiphysis and metaphysis re-gions [9] difficult. Among the other alternatives that have been explored is the CAD-based feature extraction from carpal region-of-interest (ROI) of prepubescent children and the related studies have also been positively assessed [10, 11]. However, due to the complexities surrounding limitations of the algorithm, carpal ROI has not yet been incorporated into the bone age assessment process. An interesting study for reconstruction in the field of surgical procedures was carried out by Solari et al. [12] which involves reducing postoperative CSF leak.
Deep Learning [13] and its derivatives have been successful in computer vision tasks such as ob-ject detection, classification and segmentation [14, 15]. Some valuable articles [16-18] have featured efficient means and methods for biomedical image analysis. Deep CNNs comprise pooling and convolution layers that learn hierarchical feature representations from images, followed by an ensemble of fully connected layers and dense layers that are trained on features extracted from previous layers. It has been possible to create innovative algorithms due to the availability of large datasets, most of which consist of detailed annotated features, and these algorithms/methods have increasingly boosted performances of analytical methods. Similar approaches have also been im-plemented in bone age assessment tasks [19-22], including bone segmentation for advanced feature extraction and thereby facilitating better result achievement while leaving negligible error margin rates.
In this work, two different DNN based frameworks for bone maturity estimation on the RSNA dataset constituting of 10,000 X-Ray images of the human hand are evaluated. The process involves a comparative analysis between two networks, with the base models as Inception ResNet v2 and Xception pre-trained networks. The methods suggest the superior performance of the Xception model over the Inception model, however, the Inception ResNet v2 model had a better performance during model training. The Mean Absolute Error (MAE) evaluated on the test set with the Xception model achieves best results with a deviation of around 12.583 months, whereas the Inception ResNet v2 results in a test set MAE of around 13.299 months, making the overall procedure more optimized and can thus assist in improved clinical diagnostic evaluations.
2. RELATED WORKS
The standard bone age estimation paradigm is centered around the Geurich and Pyle [3] and Tanner Whitehouse [2] methods [23]. Deep Convolutional Neural Networks [24] have been widely successful in research related to medical imaging. Pan et al. [25] applied deep transfer learning techniques such as multi-characteristic CNNs and an ensemble approach on the RSNA dataset for BAA. Their model achieved an MAE of 8.59, 6.96 and 7.35 months on all, male, and female cohorts respectively. Mansourvar et al. [26] designed an automated BAA system that used CBIR (Content Based Image Retrieval) and returned an average error rate of -0.170625 years. Rucci et al. [27] developed a scheme for bone classification using neural networks in the Tanner Whitehouse method (TW2) [2] but their results were relatively un- satisfactory with an error rate of 1.4 years. Wu et al. [28] incorporated two subnets in their deep learning based pipeline on the RSNA dataset: MASK R-CNN for eliminating background noise and a residual attention subnet based on the aforementioned subnet for generating the final predictive output and related visualizations. These techniques, however, are not well-suited for images with low resolution since they do not perform precision-based image segmentation. In a more advanced approach, Thoderg et al. [29] proposed the BoneXpert which used a repository of 3000 carefully annotated bone images and on the basis of a combination of shape, intensity and textural features, efficiently determined bone maturity. Pietka et al. [30] developed a bone age estimation method using a digital hand atlas. The preprocessing phase yielded epiphyseal/metaphyseal regions of interest (EMROIs) which there then fed to feature extraction functions. Three ratios of distance were generated: ed/md, ed/dist, and md/dist and the final assessment gave near accurate results, with only a detection failure in 4% of the radiographs. Several other such systems/methods have also been designed [31-33]. Certain algorithms [34-37] have also been established that can be applied in hand-wrist analysis, dealing with segmenting out only certain zones of the radiology images.
DCNN can be efficiently reinforced in tasks related to bone age estimation [38-40, 19]. Though some of these techniques give satisfactory results, most of them generally tend to be inclined towards some common shortcomings:
(1) The techniques might generate bias since the evaluation is centered around coarse digital processed images of hands bones.
(2) Most use regressors that are more suitable for low resolution images rather than high quality latent counterparts. This can limit the overall performance of the BAA system.
3. DEEP NEURAL NETWORKS
Multiple assessments suggest incorporation of Deep Neural Network architectures instead of Convolutional Neural Networks. Many researchers in their previous contributions have tried to use Convolutional Neural Networks for the identification of skeletal age from X-Ray images, but the methods involved using space invariant ANN’s, based on their shared-weights architecture and translation invariance characteristics. Deep Neural Networks subject to methods involving the transfer of feature maps, layer by layer as supplementary information, to perform batch-wise model preparation. Pre-trained DNN models like Inception-ResNet V2 and Xception are selected as base models, and more convolutional blocks are added to these base models to evaluate them independently.
4. METHODS
This paper proposes a method to identify the age of subjects from hand X-Ray images. This involves in- corporation of a comparison analysis of two pre-trained Deep Neural Network classifiers, namely Inception-ResNet V2 and Xception. Different evaluation parameters, such as Mean absolute error (MAE), Mean squared error (MSE), Root mean squared error (RMSE) and R-squared are used to identify the range from their predicted age and a ground truth labelled by the medical experts. The proposed method suggests the performance of models similar to medical experts and are aimed as highly useful tools for computer-aided diagnosis, towards easier age identification. The experiments were conducted using NVIDIA P100 Graphics Processing Unit (GPU). The setup further aids us to conduct experiments faster and gather results at a rapid rate than usual (Fig. 1).
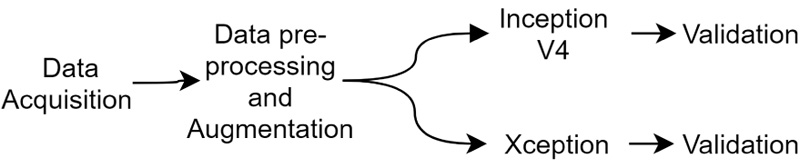
4.1. Data description
The RSNA X-Ray data has been collected from Pediatric Bone Age Challenge 2017 competition. The dataset [41] is originally contributed by Stanford University, The University of Colorado and The University of California, Los Angeles. We have taken advantage of a dataset which consists of 12,611 X-Ray images of human hand. The dataset contains hand images for image accession. The sample of X-Rays is shown in Fig. (2).

4.2. Data Pre-Processing and Augmentation
The X-Ray images were already in high-dimensional format; hence enhancing or distorting features in images was not required. However, each image was resized to 512 X 512 pixels. The images were changed to gray-scale so that the number of channels is reduced to 1, thus affecting complexity of the architectures. Image Data Generator is used to create batches of digital image data by using real-time data. This involves the augmentation of data. The data augmentation strategy in- creases data diversity for a model to increase their training capacity without any increase in training instances. This method is carried out using enhancement tools like cropping, flip, padding, resizing or changing rotation angle in order to manipulate source data.
The size of the data was reduced from 12,611 images to 10,000 images, which involves the removal of labels having duplicate or erroneous indexes. The new total is split into a training set (6,000 images), a test set (2,000 images), and a validation set (2,000 images). Upon completion of the preprocessing steps up to a sufficient standard, the DNN architectures were finally applied upon the obtained images as part of the main evaluation.
4.3. Model Architectures
The Deep Neural Network architectures used for assessment were chosen because of their optimized performance when compared to other contemporary pre-trained DNN classifiers. Both the models were initially set to train for 15 epochs. A greater number of epochs was not used since the same can prompt model overfitting, while less number of epochs can bring about an underfit model. This technique permits users to determine huge number of training epochs, training halts and determine when the model shows promising improvement across the validation dataset. Three callback techniques are utilized for model compiling, specifically ModelCheckpoint, EarlyStopping and ReduceLROnPlateau. The ModelCheckpoint callback class allows to define the location and settings to save improved model weights. The EarlyStopping callback is configured when instantiated via arguments. ReduceLROnPlateau callback monitors a parameter and if no improvement is observed for a certain number of 'patients' per epoch, the learning rate is diminished.
4.3.1. Inception ResNet v2
Inception ResNet V2 (Inception ResNet v2) [42] is a deeply convoluted neural network that is a hybrid of Inception and the ResNet modules. Here residual connections are introduced to add the output of the convolutional operations of the inception modules to the input and further the 1 X 1 convolutions are applied after the original convolutions to resemble the depth size. The residual connections replace pooling operations. The stability in the network is maintained by scaling the residual activation functions by values around 0.1 to 0.3.
As the model architecture is deep enough to overfit the data, we have employed dropout layers. These layers randomly drop out some of the nodes to bypass the complexities in the model, which in turn affects our model by losing vital patterns in the data. This problem is evaded by a layer of batch normalization. It normalizes the data to a definite range to dodge covariance shifting. The total number of parameters in the model is reduced by the global average pooling layer, thus decreasing any further chances of overfitting. The loss function is optimized by Adam optimizer [43]. The underlying equations for effective convergence and weight updates using Adam optimizer are explained in Equations 1-4.
Initial weights:
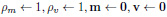 |
(1) |
Adam optimiser update equations:
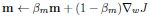 |
(2) |
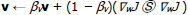 |
(3) |
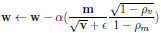 |
(4) |
Here,
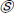 refers to element-multiplication and in Equation 4, the operations under the root are also handled element-wise.
refers to element-multiplication and in Equation 4, the operations under the root are also handled element-wise.
This model is trained upon 375 images per batch through 16 such batches during the training phase and verified on 125 images per batch for 16 batches through the validation phase. This batch size is maintained for generalization of the results. This generalization of results helps our model to predict further instances outside the training set.
4.3.2. Xception
The Xception network [44] is a deeply convoluted neural network that uses feature extraction to learn further distinct patterns in the data with a lesser estimate of parameters. The primary principle of Xception network is that it uses cross channel correlation and spatial correlation in a decoupled manner. This architecture is mainly centered around depth separable convolution accompanied by point-wise convolution, consisting of 36 convolutional layers structured into 14 modules for core feature extraction. The Xception network is the most improved form of the Inception network.
Additionally, batch normalization is used to normalize the input data, which controls the co-variance shift in the specified image data. It also enables the data to learn by itself independently. Batch normalization decreases overfitting by adding some noise in the data which enables us to use lesser dropout values. This saves the data from losing crucial visual patterns in the data. A dropout layer of 0.5 is added after the batch normalization layer. This layer is used to avoid overfitting in the data alongside keeping the crucial information in the data by batch normalization. These layers are followed by a global average pooling layer to avoid overfitting by minimizing parameters as it decreases the overall spatial dimensions of the images, reducing the model complexity for better performance. This layer is succeeded by a fully connected layer of the linear activation function to find the mean absolute error. Here Adam optimizer [43] is employed to learn and reach the global minima for optimizing the loss function. This model uses a batch size of 8, which enables us to use 750 images per training batch and 250 images per validation batch. Sequentially running these mini-batches helps in accumulating variables and updating them in succeeding batches. This helps in optimizing the memory usage and in generalizing the results by detouring from getting stuck in the local minima.
4.4. Evaluation
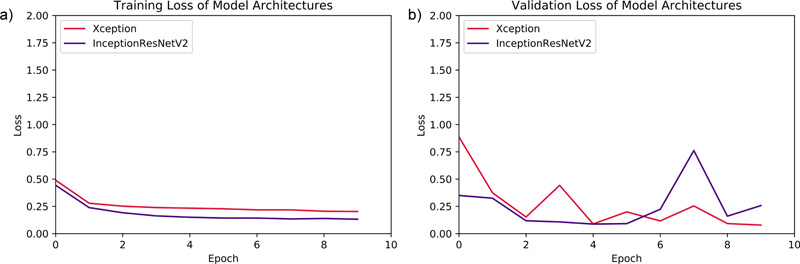
Both of the DNN models were initially set to train for 15 epochs. However, due to callbacks parameter, the Xception model was trained for 15 epochs, while Inception ResNet v2 was trained for 10 epochs and then, the training was halted. The validation-loss parameter was monitored, and mean absolute error of months was assessed for both training and validation batches per epoch. The evaluation graphs for the performance assessment of both models are demonstrated in Fig. (3).
5. RESULTS
The DNN models were compared using evaluation parameters, such as Mean Squared Error (MSE), Mean Absolute Error (MAE), Root Mean Squared Error (RMSE), and R Square value. These four error metrics are used to determine the model with more optimised results.
The Mean Absolute Error (MAE) is a metric used to determine the similarity between two sets.
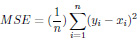 |
(5) |
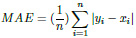 |
(6) |
The Root Mean Squared Error (RMSE) is a metric for determining the similarity between the true and predicted values. This parameter is similar to MAE, except for two conditions. Firstly, each absolute error is squared before being summed. Secondly, the final result (MSE) is square-rooted before being returned.
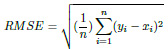 |
(7) |
The idea of R squared is that if more samples are added, the coefficient will show the prob- ability of a new point falling on the line.
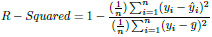 |
(8) |
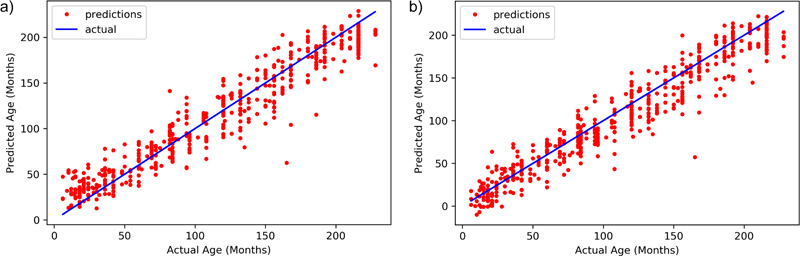
As shown in Table 1, the parameters suggest almost similar performance of both the DNN models. The performances eventually can be boosted if more data is added to the training set. Xception has a MAE of 12.583 while Inception ResNet v2 has an MAE of 13.299. The final R- Squared value is 0.943 for Xception, whereas Inception ResNet v2 achieves a value of 0.935. All these observations clearly show that Xception has a better chance of age identification than Inception ResNet v2. Fig. (4) shows the distribution of predicted results with respect to ground truth for both models.
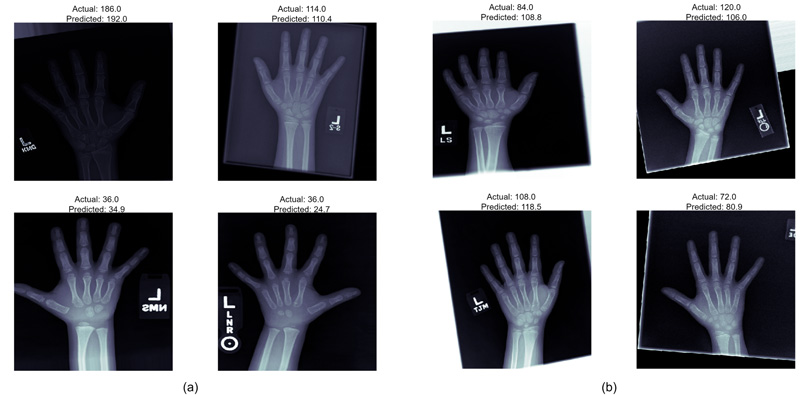
The models employed in this system were used to predict the age of the hand X-Rays in terms of months, with pre-determined ground truth set by the medical experts. The results, as demonstrated in Fig. (5), show a deviation from actual age in months in terms of MAE from the ground truth. Both the models present almost similar performances, thus guaranteeing enhanced identification examples with fewer training instances.
Additionally, both models showcase improved performances in terms of R square parameters, which denote the condition that the models already have a higher chance of correctly identifying the age if exposed to and tested upon unseen data.
| Metrics Used | Inception ResNet V2 | Xception |
|---|---|---|
| MSE | 287.328 | 254.025 |
| MAE | 13.299 | 12.583 |
| RMSE | 16.951 | 15.938 |
| R squared | 0.935 | 0.943 |
CONCLUSION
We have proposed a fully automated system for efficiently determining skeletal maturity using the RSNA dataset. The system, consisting of two primary models, Xception and Inception ResNet V2, automatically extract relevant features from the data and achieves excellent outcomes in terms of mean absolute error of 12.583 and 13.299, respectively, in the models. The results could be further enhanced if tried with system specifications allowing more reduction of the Learning Rate initially unaffected because of callbacks. The proposed model must also be exposed to more diverse training data to permit model diversity and generalization of the results, which would provide an advantageous assessment of images to make image identifications with reduced mean absolute error.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.


