RESEARCH ARTICLE
Analysis of Discrete Wavelet Transforms Variants for the Fusion of CT and MRI Images
Nishant Jain1, Arvind Yadav2, Yogesh Kumar Sariya3, Arun Balodi4, *
Article Information
Identifiers and Pagination:
Year: 2021Volume: 15
Issue: Suppl-2, M9
First Page: 204
Last Page: 212
Publisher ID: TOBEJ-15-204
DOI: 10.2174/1874120702115010204
Article History:
Received Date: 07/8/2020Revision Received Date: 24/3/2021
Acceptance Date: 01/6/2021
Electronic publication date: 31/12/2021
Collection year: 2021

open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution 4.0 International Public License (CC-BY 4.0), a copy of which is available at: https://creativecommons.org/licenses/by/4.0/legalcode. This license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Background:
Medical image fusion methods are applied to a wide assortment of medical fields, for example, computer-assisted diagnosis, telemedicine, radiation treatment, preoperative planning, and so forth. Computed Tomography (CT) is utilized to scan the bone structure, while Magnetic Resonance Imaging (MRI) is utilized to examine the soft tissues of the cerebrum. The fusion of the images obtained from the two modalities helps radiologists diagnose the abnormalities in the brain and localize the position of the abnormality concerning the bone.
Methods:
Multimodal medical image fusion procedure contributes to the decrease of information vulnerability and improves the clinical diagnosis exactness. The motive is to protect salient features from multiple source images to produce an upgraded fused image. The CT-MRI image fusion study made it conceivable to analyze the two modalities straightforwardly.
Several states of the art techniques are available for the fusion of CT & MRI images. The discrete wavelet transform (DWT) is one of the widely used transformation techniques for the fusion of images. However, the efficacy of utilization of the variants of wavelet filters for the decomposition of the images, which may improve the image fusion quality, has not been studied in detail. Therefore the objective of this study is to assess the utility of wavelet families for the fusion of CT and MRI images. In this paper investigation on the efficacy of 8 wavelet families (120 family members) on the visual quality of the fused CT & MRI image has been performed. Further, to strengthen the quality of the fused image, two quantitative performance evaluation parameters, namely classical and gradient information, have been calculated.
Results:
Experimental results demonstrate that amongst the 120 wavelet family members (8 wavelet families), db1, rbio1.1, and Haar wavelets have outperformed other wavelet family members in both qualitative and quantitative analysis.
Conclusion:
Quantitative and qualitative analysis shows that the fused image may help radiologists diagnose the abnormalities in the brain and localize the position of the abnormality concerning the bone more easily. For further improvement in the fused results, methods based on deep learning may be tested in the future.
1. INTRODUCTION
Image fusion has been employed in a variety of applications, viz. remote sensing, medical imaging, military, and astronomy. The bits and pieces provided by different imaging modalities can be knitted for a better understanding of the entity under observations. For a faithful replication of the human organ with or without pathology, such an idea is appreciated. Several image registration and fusion techniques have been discussed in the context of applying them to patient studies obtained during clinical workup. Wavelet transformation-based methods are explored extensively for multimodal image fusion, but an exhaustive comparison of various mother wavelets under discrete wavelet transform is not documented yet.
The determination of the imaging methodology for a medical investigation requires clinical bits of knowledge explicit to organs under examination. It is difficult to catch all the subtleties from one imaging methodology that would guarantee the clinical exactness and strength of the investigation and resulting diagnosis. The intuitive idea is to take a gander at images from different modalities to make a faithful and precise evaluation. A trained medical practitioner can evaluate subtleties that supplement the individual modalities. For instance, Computed Tomography is broadly utilized for tumour and anatomical identification, though soft tissues are better captured by magnetic resonance imaging. Further, T1 weighted MRI gives insights concerning the anatomical structure of tissues, though T2 weighted MRI highlights normal and pathological tissues [1].
The fusion of the images obtained from these two modalities that is CT and MRI, to form a single fused image, will help radiologists diagnose the abnormalities in the brain and localize the position of the abnormality concerning the bone. The motive is to protect salient features from multiple source images to produce an upgraded fused image. The CT-MRI image fusion study made it conceivable to analyze the two modalities straightforwardly. Multimodal medical image fusion procedure contributes to the decrease of information vulnerability and improves the clinical diagnosis exactness.
A comprehensive study of multisensor image fusion based on wavelet was executed by H Li and team, where the emphasis was on fusion rule [2]. They have experimentally evaluated the performances of simple fusion rule versus area-based maximum selection rule and demonstrated the effective extraction of salient features at different resolutions. David A. Yocky [3] also tested a wavelet merger for the fusion of panchromatic and spectral images and compared the findings with the fusion results of the intensity-hue-saturation (IHS) merger.
In the quest of finding the best schema of image fusion, hybridization of existing techniques and updating of fusion rules had been devised. A regular DWT was made more skilled for multimodality fusion by Yufeng and his team [4] by adding the power of Principal component analysis (PCA) and morphological processing, and they called it advanced DWT (aDWT). They also suggested a new quantitative metric, “ratio of spatial frequency error” for fusion assessment. The hybridization of various techniques for the betterment of fusion results is one of the most experimented methods [5–9].
An unorthodox fusion scheme was proposed by Yong Yang et al. [10], where they exploited maximum absolute values and maximum variance rules for low frequency and high-frequency coefficients, respectively. There is a study [11] where authors have used wavelet transform modulus maxima rather than wavelet transform coefficients and claimed that in this manner, the edge and margin information could be preserved.
The MRI-PET (Positron emission tomography) image pair is among the fairly explored multimodal fusion experimentation. A 4-level DWT was employed by Bhavna et al. [12] to synthesize PET images into its high activity and low activity regions, which were further fused with MRI images in the transformed domain. The authors concluded that their DWT-based fusion method outperforms the conventional fusion techniques. In a row of applying multilevel DWT, there is one such study where authors have compared DWT versus 3-level DWT on CT-MRI pairs and demonstrated the superiority of multilevel based on visual as well as some quantitative parameters.
Siddalingesh et al. [13] have declared the superiority of feature-level image fusion, although it takes much time. They have compared DWT, stationary wavelet transforms (SWT), and dual-tree- complex wavelet transform (DT-CWT) and concluded DT-CWT as the winner as it takes care of directionality. The compendium of multimodality fusion is full of experimentation with hybridization, updating infusion rules, new versions of the wavelet transform, and deep neural networks. However, an exhaustive study of DWT families is not conducted.
The rest of the paper is structured as follows: Section-2 presents the methodology employed for the investigation of the efficacy of the wavelet functions for the fusion of the CT & MRI image. The result and discussion section are detailed in section-3. The work has been concluded in section-3.
2. METHODOLOGY
The schematic representation of the proposed approach for the investigation of the suitability of DWT wavelet families for image fusion is illustrated in Fig. (1).
The essential requirement for the CT & MRI images is that they should be spatially registered [14]. These input images are individually transformed into wavelet domains with the help of forwarding discrete wavelet transform (FDWT). For the current investigation, we have restricted the image decomposition until level-2 only. Wavelet family’s viz., Haar, Daubechies, Coiflet, Biorthogonal, Reverse biorthogonal, Symlets, Discrete approximation of Meyer wavelet, and Fejer-Korovkin wavelets have been used for decomposition. A total of 120 variants of wavelet family members have been employed here, and the efficacy of individual wavelet functions on the fusion result has been investigated. It has been established that simultaneous localization in spatial and frequency domain is accomplished using wavelet transform. Thus, the rationale behind the use of the max fusion rule only is justified with the help of the following two cases.
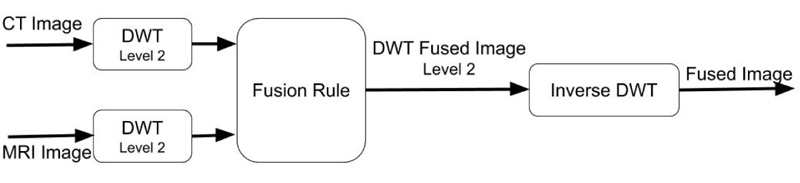 |
Fig (1). Flowchart of the proposed method. |
Case 1: When the object of interest looks quite distinctive in image 1 in contrast to image 2, the fused image obtained with the max rule will preserve the object of interest in image 1, whereas the same object in image 2 will be discarded.
Case 2: Let the outer boundary and inner boundary of image 1 and image 2 look distinctive from each other; under such a scenario, the wavelet coefficients of both the images will be distinctive at different scales. When applied max fuse rule, it can retain the distinctive information of image 1 and image 2, respectively, in the fused image.
The fused image in the wavelet domain is transformed into the spatial domain using an inverse discrete wavelet transform.
3. THE THEORETICAL BACKGROUND OF DISCRETE WAVELET TRANSFORMS
The most standard wavelet decomposition utilized for image filtering is DWT [15]. The Wavelets are mathematical functions used for decomposing the signal into various frequency components and, after that, concentrate every part with a resolution coordinated to its scale. In two-dimensional DWT, the one-dimensional DWT is applied along the rows in the first step, and then the columns of the result are processed to produce four sub-images in the transformed space: LL, LH, HL, and HH. The DWT is widely used in various image handling applications, for example, image denoising, segmentation, feature extraction [15, 16], and image indexing [17, 18]. Fig. (2) illustrates the level 1 DWT decomposition of an image.
In this paper, eight types of wavelet filters (Haar, Daubechies, Symlet, Coiflet, Fejer-Korovkin, DMeyer, Biorthogonal, and Reverse Biorthogonal) have been used. Haar wavelet is the very first and simplest wavelet family that belongs to the Finite Impulse Response (FIR) filter. It is discontinuous and bears a resemblance to a step function [19]. Daubechies is a compactly supported orthonormal wavelet family that was proposed by Ingrid Daubechies. The variants of the Daubechies family are represented as dbN, where “db” corresponds to the surname of the inventor and N is the order. The db1 wavelet bears characteristics similar to the Haar wavelet [19, 20]. The Bioorthogonal wavelets are symmetric and found to have compact support [21]. It makes use of two wavelets, one for decomposition and the second one for reconstruction to extract useful properties. This wavelet is most suitable for the reconstruction of signals and images as it exhibits linear phase property.
At the request of Ronald Coifman, Ingrid Daubechies has designed another wavelet called Coiflets which is nearly symmetrical as well as features scaling functions with vanishing moments. Further, Daubechies has proposed Symlets, which has nearly symmetrical wavelet characteristics obtained by modifying the db family [22].
Fejer-korovkin is indeed symmetrical and soft as opposed to db. Approximation theory is the area where it finds major applications. A seminal contribution in the field of wavelet filtering was made by a harmonic analyst, whose FIR-based approximation was called Discrete Meyer (DMeyer), which possesses infinite differentiable properties. It is an orthogonal, symmetric, and compactly supported wavelet. To acquire more freedom than conventional orthogonal wavelets, biorthogonal wavelets were introduced, which must be invertible. It uses two wavelets for decomposition and reconstruction. With the use of biorthogonal spline wavelets, the Reverse Biorthogonal wavelets were produced, which preserves the symmetrical nature of the filter and reconstruction.
4. RESULTS & DISCUSSION
In this section, the efficacy of the wavelet families on the fusion of registered CT & MRI images has been investigated. All the results are obtained using the MATLAB 2016b version installed in 2.4 GHz CPU, Core i5, 4 GB RAM, 128 GB SSD, and Windows 10 Operating system. The dataset and performance evaluation matrix is described in the following subsections:
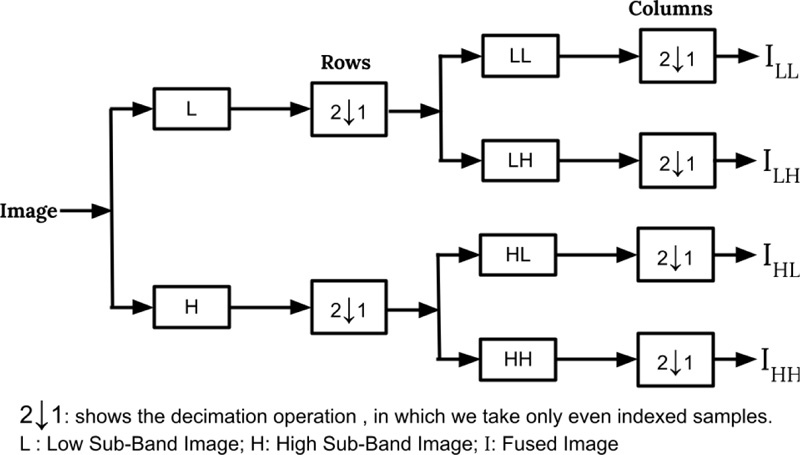 |
Fig (2). Block diagram representing first level DWT decomposition for an image. |
4.1. Dataset
To investigate the performance of 120 wavelet family members, three datasets of pairs of CT and MRI images are collected from an online database available at http://www.med.harvard.edu. The pair of CT and MRI images are already registered with each other, due to which image fusion methods are directly implemented on these datasets. Dataset 1 is a case of metastatic bronchogenic carcinoma and belongs to a 42-year-old woman. Dataset 2 belongs to a patient of 63 years of age suffering from acute stroke, who can write but cannot read. Dataset 3 is a case of a 22-year-old patient suffering from Sarcoma. Several brain slices were obtained with the complete scan of the brain of all the patients. All the brain slices are in the axial plane. Slice numbers 10, 12, and 20 are randomly selected for testing the performance of different wavelet families. All the images of the datasets are in grayscale format and are of the size 256×256.
4.2. Performance Evaluation Matrix
The image fusion results can be evaluated using subjective and objective approaches. The subjective approaches are considered to be complex as they are based on psychovisual testing and cannot be applied in an automatic system. In addition to this, the inter-observer variability is the challenge in image fusion, which leads to the need for a quantitative parameter evaluation matrix to measure the efficacy of fused images. The present work employs classical evaluation parameters and gradient information parameters. Classical evaluation parameters used in this work are correlation (average normalized correlation), mutual information (MI), entropy, fusion symmetry (FS), spatial frequency (SF), and average gradient (AG). The values of these parameters should be higher for better fusion. The gradient information [23] parameters considered here are QAB/F, LAB/F, NAB/F, which stands for the total amount of information transferred from source image to fused image, total loss of information, and artifacts/noise added to fused image during the fusion process, respectively. Out of these three parameters, a higher value of QAB/F, whereas lower values of LAB/F and NAB/F are expected.
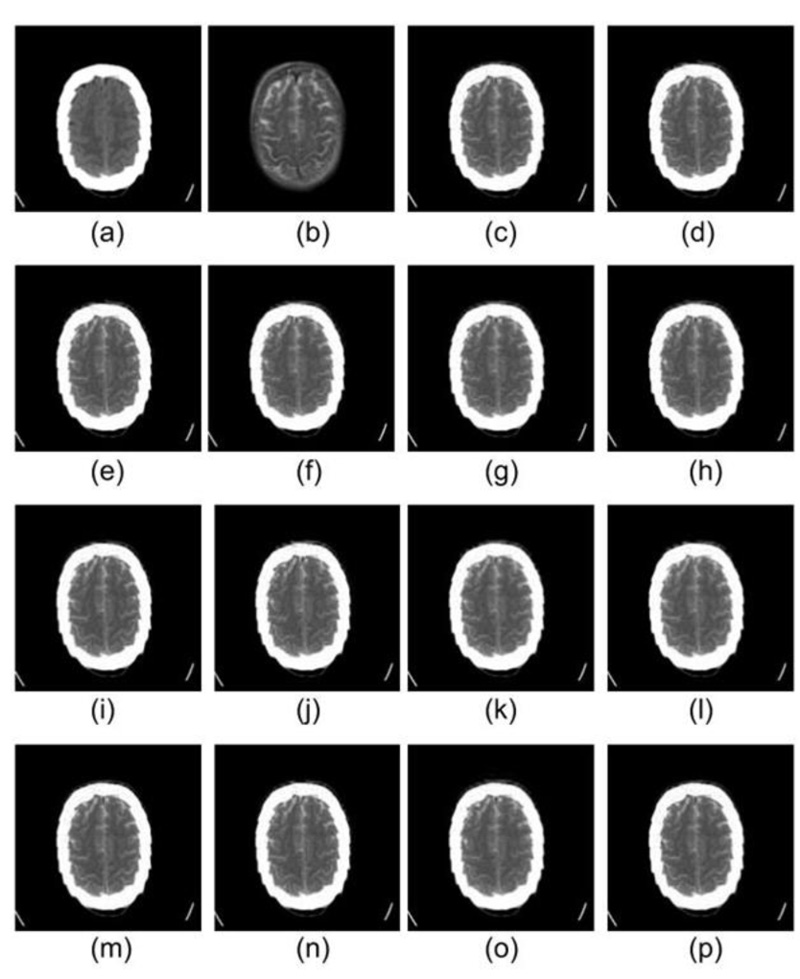
The images under investigation have been first processed using DWT, which produces four sub-images at the 1st level of decomposition, namely LL, HL, LH, and HH. In the second level of decomposition, the LL sub-image of the 1st level of decomposition has been further decomposed to obtain 4 images (LL, HL, LH and HH). Subsequently, the max fusion rule has been applied to the decomposed sub-images (i.e., 7 sub-images obtained from the decomposition). The fused image is then transformed back into a spatial domain using inverse discrete wavelet transform (IDWT), which shall be useful to the doctors or can be used by the machine for further analysis. The visual representation of the images produced at every stage is shown in Fig. (3).
All the 120 wavelet filters are exhaustively tested on all three datasets. Out of 120 wavelet family members, those who have produced better-fused images in terms of visualization are as follows: Haar, db1 & db2 (Daubechies wavelet family), sym2 & sym4 (Symlets wavelet family), coif2 & Coif4 (Coiflet wavelet family), fk4 & fk6 (Fejer-Korovkin wavelet family), dmey (DMeyer wavelet family), bior2.2 & bior2.4 (Biorthogonal wavelet family), and rbio 1.1 & rbio1.3 (Reverse Biorthogonal wavelet family). The top two results of all the eight families in terms of both visual appearance as well as quantitative parameters are displayed here.
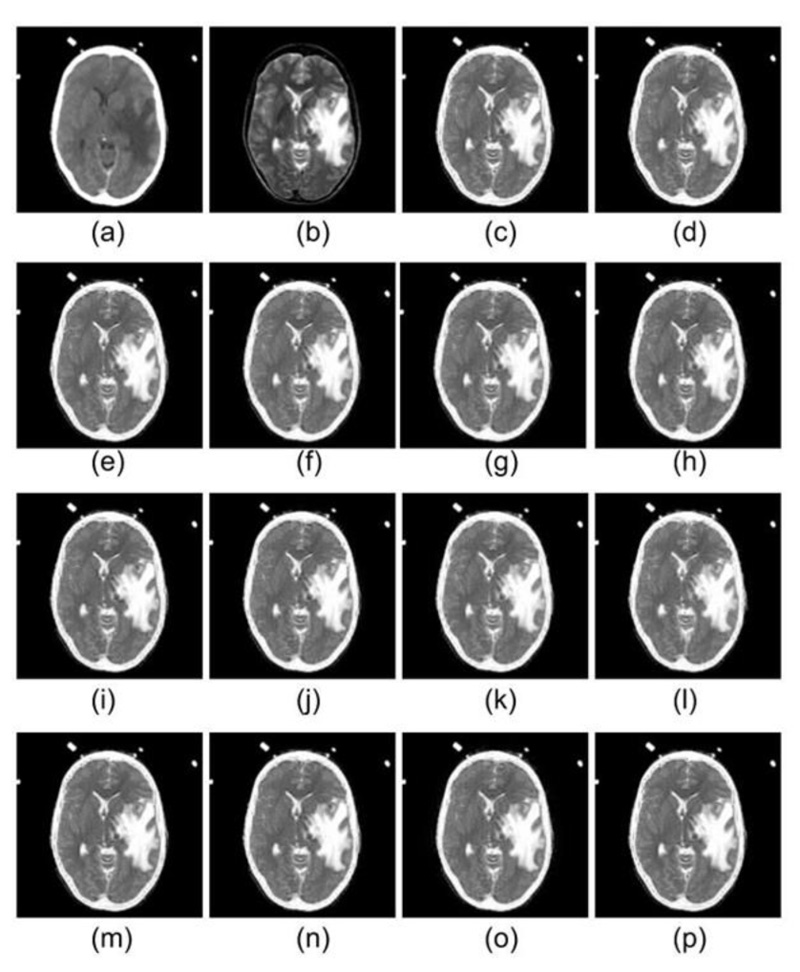
Figs. (2-6) represent the input dataset used and the corresponding fused images obtained using the following wavelets: db1, db2, sym2, sym4, coif2, coif4, fk4, fk6, bior 2.2, bior 2.4, rbio 1.1, rbio1.3, dmey, and Haar. In Fig. (3), the input CT and MRI images from dataset 1, which are fused using different wavelet family members, are displayed in 4(a) and 4(b), respectively. Since soft tissues of the brain are best scanned with MRI, whereas bone information is collected with CT, therefore the best fusion method should preserve information of the soft tissues from MRI, whereas information of the bones must be taken from CT image. Analyzing the fused images represented in Fig. (4), it has been observed that the texture of the soft tissues present in MRI got mixed with the information of the soft tissue in CT, due to which texture loss in the soft tissues is visible in all the fused images obtained with different wavelets. However, comparing all the fused images with each other, it can be seen that the texture of the soft tissues is somewhat better visible in the fused images obtained from db1, rbio 1.1, and Haar wavelets.
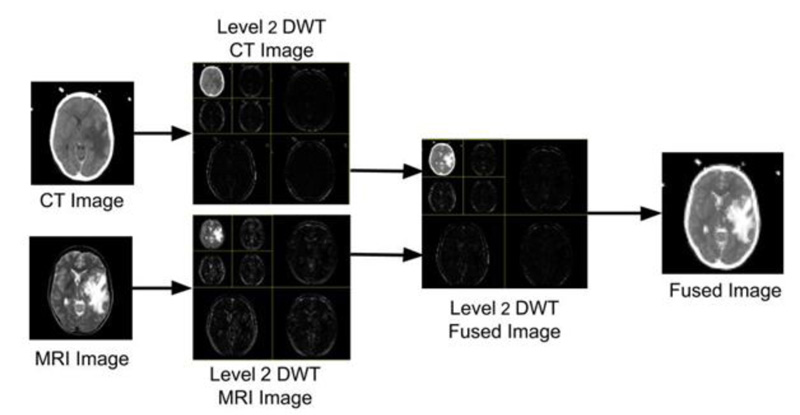 |
Fig. (3). Fusion of CT & MRI images using 2nd level DWT and max rule-based fusion. |
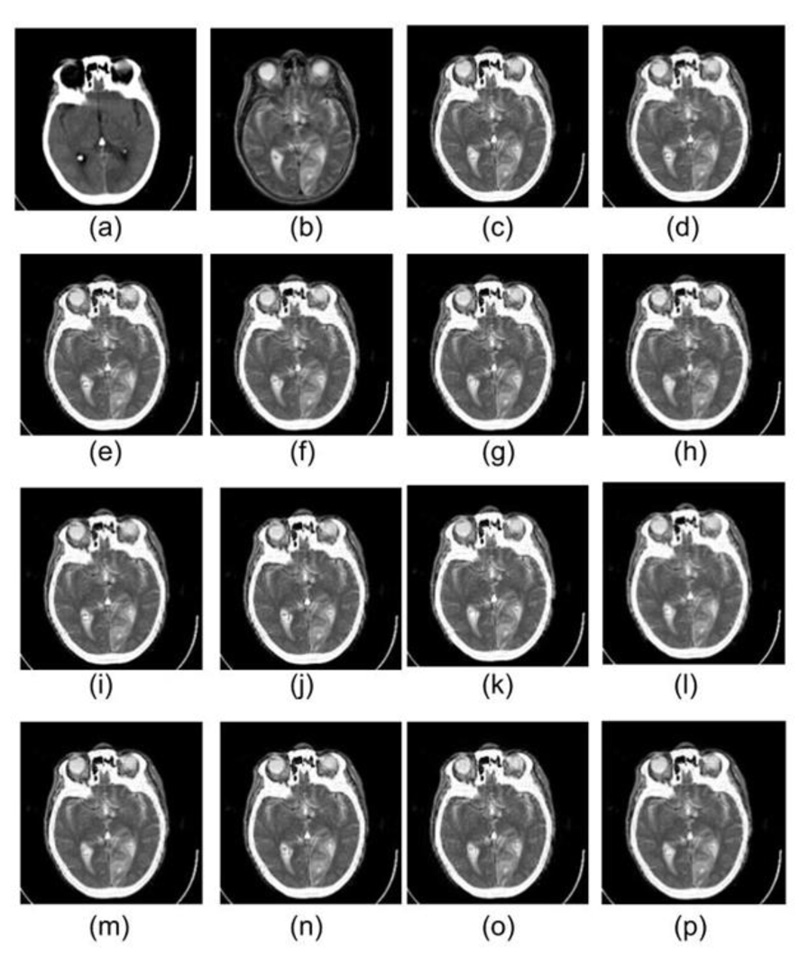
Dataset 1 consists of MRI and CT scans having metastatic bronchogenic carcinoma, which is visible in the MRI scan compared to the CT scan. It should appear the same in the fused images as it appears in the MRI. It has been observed that metastatic bronchogenic carcinoma is visible in all the fused images, as illustrated in Fig. (4). However, since the soft tissue texture is visible in a better way from the fused images obtained using db1, rbio 1.1, and Haar wavelets, therefore the contrast between the soft tissue and the carcinoma is better in the three fused images as compared to fused images obtained from other wavelets.
As it comes to the bone structure information present in the fused images, it has been observed that some information loss took place in the fused images. Although bone appears good in CT, the transition from the skull to the brain is smoother as compared to the MRI scans. Thus the boundary of the skull is pretty visible in the MRI. Consequently, when the fusion rule is applied, it emphasizes the edges of MRI, and it is evident in the results of all the wavelet family members.
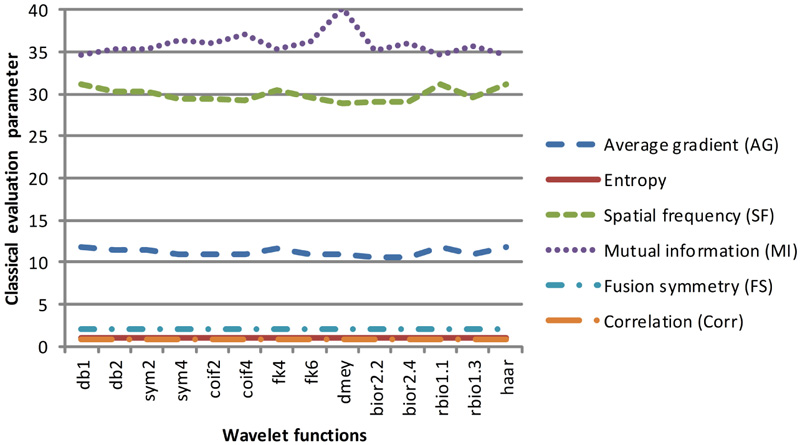 |
Fig (7a). Classical evaluation parameters of dataset 1 for selected wavelet families. |
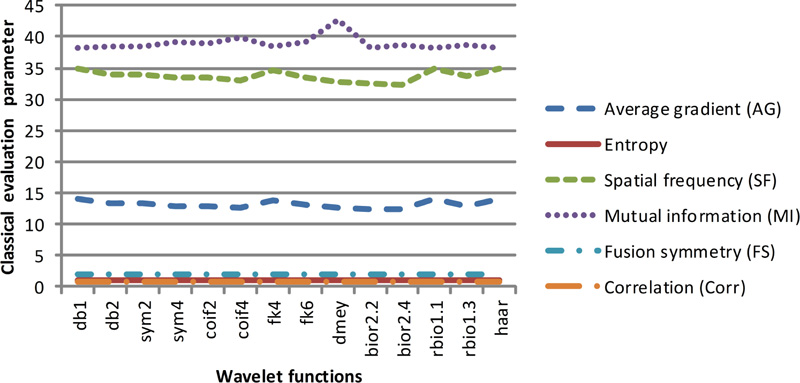 |
Fig (7b). Classical evaluation parameters of dataset 2 for selected wavelet families. |
Similarly, in Figs. (5 and 6), images (a) and (b) represent CT & MRI input images of datasets 2 and 3, respectively. The remaining images in both the figures are the fused images obtained by the application of selected wavelets families. For these two datasets also db1, rbio1.1, and Haar wavelet have produced better visual quality fused images, which are in line with dataset 1 discussed earlier in detail.
In the light of the above discussion, it can be concluded that fused results obtained from db1, rbio 1.1, and Haar wavelets are much better in comparison to other wavelets from a qualitative analysis perspective. But as subjective differs from person to person, to prove the efficacy of the fusion image quality, quantitative performance analysis needs to be performed from the machine vision perspective. Therefore, two performance measurement classes, namely, classical quantitative analysis and gradient information parameters, have been calculated for the fused images.
These two classes of quantitative parameters have been illustrated in Figs. (7a-7d), respectively. As per the literature, the six classical qualitative parameters should be higher, but for the fused images obtained using wavelet filters db1, rbio 1.1, and Haar, the value of AG and FS1 is high. In contrast, the value of entropy, FS, and MI are low. Altogether the values of classical parameters are not in harmony with the visual appearance. Thus, their validity for the justification of fusion results cannot be trusted.
The gradient-based quantitative analysis represented in Fig. (8a-8d) depicts that the value of QABF is on the higher side. In contrast, LABF and NABF are on the lower side for wavelet filters db1, rbio 1.1, and Haar (The summation of the three parameters should be equal to unity. If the value is more than unity, then NABF1 is calculated). This goes directly in favor of the visual analysis. Hence, it can be concluded that gradient-based quantitative parameters are faithful parameters to determine the efficiency of the fusion method for MRI-CT.
With quantitative analysis, it is proved that the fused image carries all the relevant information from both CT and MRI images, whereas qualitative analysis shows that the fused image carries all the visible information from both CT and MRI images, therefore fused image may help radiologists for diagnosing the abnormalities in the brain and in localizing the position of the abnormality concerning the bone more easily.
The DWT-based CT-MRI image fusion method presented in the paper is feasible for the design, optimization, and verification of treatment planning in the context of making appropriate and precise decisions by medical experts. Further, previous feasibility studies [24, 25] of medical image fusion have also suggested that the treatment planning based on fused CT and MRI images resulted in improved target volume and risk structure definition.
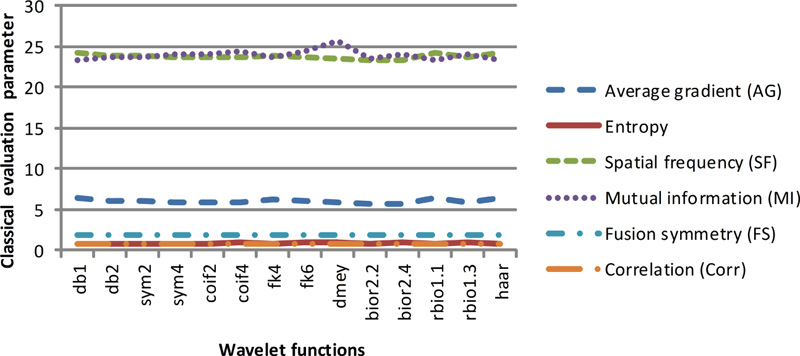 |
Fig 7(c). Classical evaluation parameters of dataset 3 for selected wavelet families. |
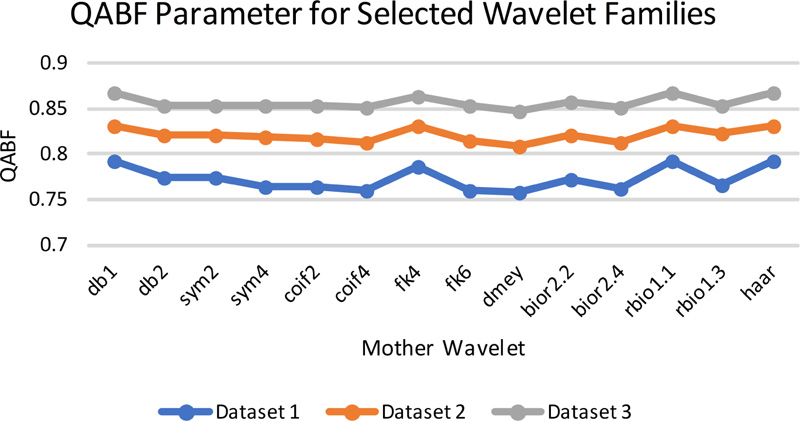 |
Fig 8(a). QAB/F evaluation parameters for selected wavelet families. |
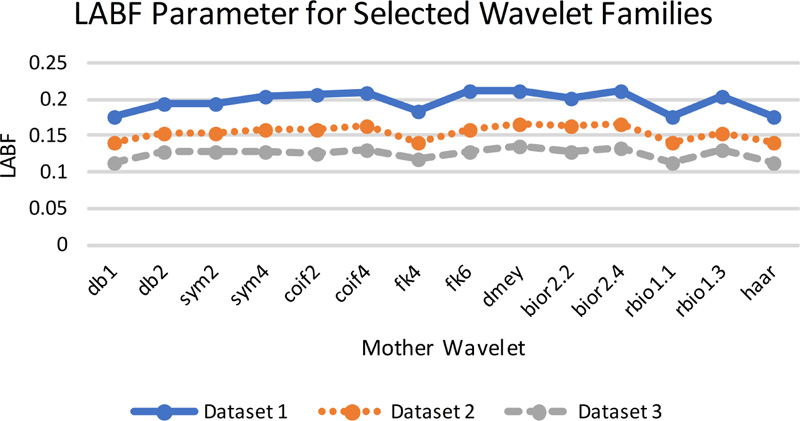 |
Fig (8b). LAB/F evaluation parameters for selected wavelet families. |
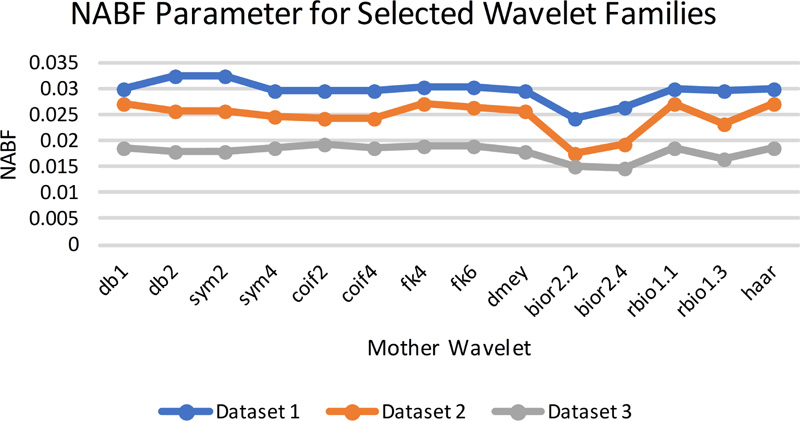 |
Fig (8c). NAB/F evaluation parameters for selected wavelet families. |
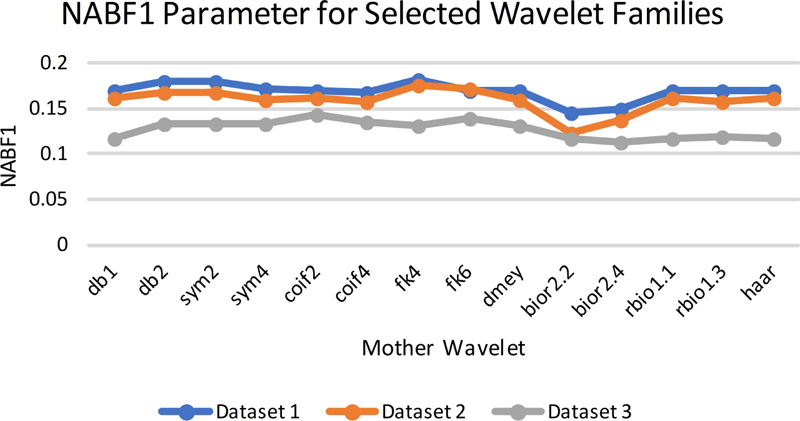 |
Fig (8d). NAB/F1 evaluation parameters for selected wavelet families. |
CONCLUSION
In this work, the authors have investigated the influence of eight different wavelet family members on the image fusion quality for registered CT & MRI images. The max fusion rule has been used for investigating the efficacy of the DWT family members for improving the fused image quality. Out of the 8 family members under the study, it is observed that db1, rbio 1.1, and Haar wavelet functions have given the best fusion result in the context of the visual quality of the fused images. Further, to strengthen our observation in addition to the subjective approach, the classical and gradient information evaluation parameters have also been calculated. It is observed that the gradient information evaluation parameters are supporting the visual appearance of infused images. However, the classical evaluation parameters are not in line with the visual appearance of the fused images.
Quantitative and qualitative analysis shows that the fused image may help radiologists diagnose the abnormalities in the brain and localize the position of the abnormality concerning the bone more easily. For further improvement in the fused results, methods based on deep learning may be tested in the future.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data set is available in the "The Whole Brain Atlas" at: https://www.med.harvard.edu/aanlib/. Further details of the dataset are given in the dataset link itself.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared None.