All published articles of this journal are available on ScienceDirect.
Multi-Channel Local Binary Pattern Guided Convolutional Neural Network for Breast Cancer Classification
Abstract
Background:
The advancement in convolutional neural network (CNN) has reduced the burden of experts using the computer-aided diagnosis of human breast cancer. However, most CNN networks use spatial features only. The inherent texture structure present in histopathological images plays an important role in distinguishing malignant tissues. This paper proposes an alternate CNN network that integrates Local Binary Pattern (LBP) based texture information with CNN features.
Methods:
The study propagates that LBP provides the most robust rotation, and translation-invariant features in comparison with other texture feature extractors. Therefore, a formulation of LBP in context of convolution operation is presented and used in the proposed CNN network. A non-trainable fixed set binary convolutional filters representing LBP features are combined with trainable convolution filters to approximate the response of the convolution layer. A CNN architecture guided by LBP features is used to classify the histopathological images.
Result:
The network is trained using BreKHis datasets. The use of a fixed set of LBP filters reduces the burden of CNN by minimizing training parameters by a factor of 9. This makes it suitable for the environment with fewer resources. The proposed network obtained 96.46% of maximum accuracy with 98.51% AUC and 97% F1-score.
Conclusion:
LBP based texture information plays a vital role in cancer image classification. A multi-channel LBP futures fusion is used in the CNN network. The experiment results propagate that the new structure of LBP-guided CNN requires fewer training parameters preserving the capability of the CNN network’s classification accuracy.
1. INTRODUCTION
Breast cancer is the most prevalent form of cancer in women, comprising 14 percent of Indian women's cancers. In both rural and urban India, breast cancer seems to be on the rise. A Breast Cancer Statistics 2018 survey [1] revealed a total of 1,62,468 new registered cases, and 87,090 reported deaths in a year. In higher stages of development, the survival of cancer is challenging, with over 50 percent of Indian women living with stage 3 and 4 breast cancer. Post-cancer survival was registered at 60% for breast cancer women in comparison with the U.S. having 80%. The biopsy is a diagnostic procedure commonly used to collect tissue samples of a human subject and analyze the existence or extent of a disease by a pathologist using a microscope. For observation, these tissues are processed and stained. Optical coherence tomography (OCT) provides an innovative non-invasive modality of optical imaging that can provide three-dimensional, high-resolution images of biological tissue structures omitting the requirement of the staining process [2]. The classification of breast tissue types with optical coherence microscopy (OCM) can be improved further by incorporating texture analysis. The combination of OCT and confocal microscopy have a better depth of penetration than confocal microscopy and higher resolution than OCT [3].
Zhou et al. [4] developed an integrated system of OCT and OCM in their laboratory. They presented a comparative analysis of corresponding histologic sections with this integrated imaging for the identification of distinguishing features or characteristics to differentiate benign and malignant breast lesions at various resolutions. Fig. (1) shows the features observed by the authors in normal breast tissue and malignant tissue for both OCM/OCT images and stained images.
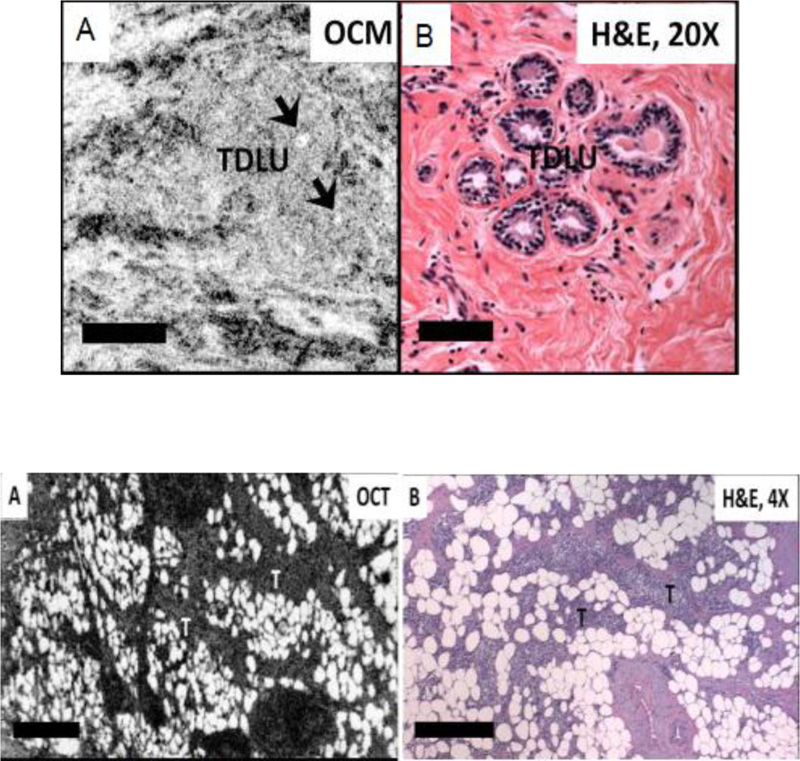
Fig. (1) shows that the apparent curved hypo scattering monomorphous circular structures in adipose tissue using OCT/OCM image is clearly visible in normal tissue. The next image in Fig. (1) shows the observed feature in benign tissue in which different sizes of necrotic (hyposcattering) adipocytes are visible clearly in the OCT image. This work demonstrates that texture pattern plays an important role in tissue classification.
Texture analysis has been used in many biomedical applications [5-7]. This texture analysis method can be categorized into four classes: (a) statistical approaches, (b) structural approaches, (c) Model approaches, and (d) Transformation approaches. Statistical approaches include a Gray level co-occurrence matrix (GLCM), local binary pattern (LBP), auto-correlation function (ACF), and histograms feature [8]. The morphological operations [9], primitive measurements [10], and skeleton representation [11] are the subpart of the structure-based texture analysis. The auto-regressive model and random field generation [12] are the subclasses of model-based texture representation. Wavelet transform [13], Gabor transform [14], curvelet transform [15] etc analyzes the texture information using transformation methods. Statistical approaches are versatile in contrast to structure-based strategies. They do not include pictures that include repeated patterns of form and are better adapted to the study of OCT / OCM tissue specimens.
More recently, the use of a hierarchical training framework in deep learning is the most robust approach for feature extraction and classification in computer vision applications. Many deep learning approaches have been used in the human breast tissue classification yielding excellent performance, which is further discussed in Section 2.
A local binary pattern having robust discrimination capabilities can reduce the burden of CNN and can enhance the performance of CNN for better classification [16]. Therefore, this work represents the breast cancer image classification using the integrated approach of texture pattern analysis and deep learning. To take advantage of both spectral and spatial features, a deep CNN architecture using local binary pattern-based texture features is proposed in this paper. We followed the algorithm proposed in earlier work [17]. The CNN architecture is trained from scratch using the BreaKHis dataset [18] for binary classification. LBP generally works with gray-scale images. This approach offers the advantages of significant parameter saving, fast convergence, and avoids overfitting problems.
The overall structure of the paper is as follows: Section 2 introduces works of literature on histopathological image classification. The survey is divided into two parts: (a) texture features based classification models and (b) various deep learning approaches used in the breast cancer image classification. Section -3 introduces LBP and CNN structure to use these LBP features. Experimental analysis and results are discussed in section 4. At last, concluding remarks are presented in section 5.
2. RELATED WORK
Histopathology images demonstrate the effect of the influence of diseases through the underline tissue structure, which is usually preserved during staining image processing. The structure analysis is a crucial step in the diagnosis process in the detection and classification of these histopathological images. Previously, segmentation approaches were used for the classification of images [18]. Later region of interest was identified using active contour formulation [19]. The technical advancement in computer-based vision processing leads to various alternates in the analysis of histopathological images. The wavelet transform based texture analysis was used [20, 21] to detect lung cancer. Gabor filter provides texture characterization at various scales, and orientation also offers good advantages in breast cancer detection and classification [22]. GLCM based texture classification was also demonstrated for prostate and skin cancer detection [23]. A local structure tensor was also used to extract the texture information and image segmentation [24]. Vast CNN models are presented in the literature. However, we limit our study to LBP and LBP integrated CNN networks. This section initially presents the use of local binary pattern in cancer image classification, and later use of CNN and LBP in histopathological images are presented.
2.1. LBP in Histopathological Image Classification
LBP is largely utilized in computer vision applications to describe image features since 1994. LBP was used effectively in the texture pattern classification in MRI brain images [25]. These features were used for the content-based image retrieval process. Pereira et al. [26], used LBP features in the support vector machine (SVM) classification model and tested the algorithm on the Mammographic Image Analysis Society (MIAS) dataset. Pawar et al. [27] enhanced the contrast of mammogram images using local entropy methods and segmented the RoI from the images. To reduce the false diagnosis, LBP features were used along with curvelet transform. Later NN was used for the classification. Öztürka and Akdemir compared well-known texture feature extractors, i.e., GLCM, LBP, Local Binary Gray Level Cooccurrence Matrix (LBGLCM), grey-level run-length matrix (GLRLM), and Segmentation Based Fractal Texture Analysis (SFTA) [28] and concluded that LBP is more robust amongst all texture feature operators achieving highest accuracy in texture image classification.
A mass detection and mammographic image classification using curvelet and LBP were proposed by Tosin et al. [29]. K-nearest neighborhood (KNN) was used in the classification achieving 96% accuracy on the MIAS dataset. Farhan and Kamil segmented MIAS images and obtained LBP features from the region of interest [30]. Using logistic regression classifier, they received 85.5% classification accuracy. Authors previously concluded [31] that LBP and its various variants are competitive enough to extract robust features for breast density classification. They achieved 92% accuracy with extended LBP and Bayesian networks in comparison with KNN, SVM, and Random forest classification algorithms.
Kulshreshtha et al. [32] proposed content-based mammogram image retrieval. They cropped the mammogram images, extracted features from the cropped images, and applied k-mean clustering. Then, a query image was matched with clustered images to retrieve it. A study on the role of various texture feature operators for tissue region classification was carried out by Kather et al. [33]. For further improvement in recognition, they also combined multiple texture features using LBP, GLCM, and Gabor operators. Simon et al. adopted the LBP features over a three-color plane for Glomerular Detection from histopathology image using SVM. They achieved an overall 97.5% precision score in the detection.
From the study, it has been analyzed that amongst all texture feature operators, LBP features provide the most robust features. The LBP is mainly applied to extract the features from mammogram images, and very few algorithms were proposed where LBP was used with histopathological images.
2.2. Histopathological Image Classification using LBP and CNN
A comparative analysis between CNN, LBP, and Bag of Visual word (BOVW) was presented by Kumar et al. [34]. They tested each model on the KIMIA Path960 histopathological image dataset containing 20 different classes of a total of 960 histopathology images. Stand along LBP achieved 90.62% accuracy whereas, the dictionary approach using LBP features obtained 96.50% accuracy. Due to the small size dataset, CNN's accuracy was limited to 94.72%. Mojahead et al. [35] presented a CNN network for Breast Margin Assessment from OCT images. A high axial resolution of 2.7 µmm and 5.5 µmm lateral resolution images were used in the study. They obtained an 84% F1 score in tissue classification. A transfer learning approach of CNN was used to access the mammographic breast density [36]. The comparative analysis shown in this paper concluded that CNN outperformed in comparison with local and global feature extracted method with 84.2% accuracy. Majid Nawaz et al. classified histopathological images in four classes using deep CNN [37]. They modify the DenseNet network by adding four dense layers and three transition layer and obtained 95.4% classification accuracy. The authors used Haar wavelet [38] for texture features, and these features were integrated into the CNN network for image classification problems. They obtained 96.85% accuracy on the BreaKHis dataset. In another work [39], the authors modified the CNN architecture by concatenating spectral features from the wavelet transform with spatial features obtained using convolution operation. Many different varieties of CNN architecture, which totally depend on the spatial feature using convolution operation of CNN for histopathological image classification, were proposed in the literature [40-42]. Though texture structure plays an equivalent role in histopathological images, it has been dominated or not fully used in the CNN architecture. Therefore, this paper presents an alternate structure of CNN that fully utilizes the LBP based texture structure information along with spatial information of CNN.
3. METHODOLOGY
As discussed above, most existing algorithms have used either the deep CNN (DNN) algorithm or handcrafted texture features in the detection and classification model. Individually both models have shown promising results. LBP has provided competitive results to deep CNN-based methods, particularly in the presence of rotations and noises [43]. However, to reduce the burden of CNN while maintaining the performance of the system, we presented an alternate way of fusion of the handcrafted texture features with deep learning architectures.
The local binary pattern is a powerful operator to discriminate texture features among all texture feature operators. It provides rotation and translation invariant features. In an image plane I(x,y), the LBP operator finds a similarity between the neighbor pixels over predefined block size. Let (Xc, Yc) be the center of the block with the size PxP. Then LBP is obtained by comparing center pixels with its P2-1 neighbor pixels residing in the circle of radius R. It can be expressed mathematically as,
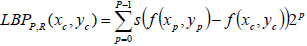 |
(1) |
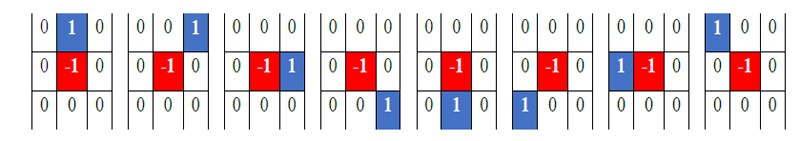
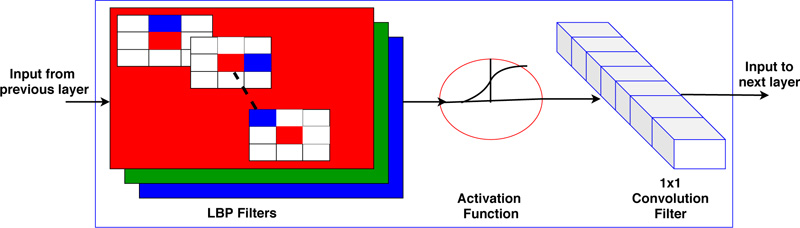
Where s (·) = 1 if f (xp, yp ) ≥ f (xc, yc ) otherwise s (·) = 0. To generalize the description of LBP, few variations are as follows: LBP is generally encoded using a base of two. Thus weights assigned to each bit are guarded to the power of two. It can be generalized by allowing to take any real value by the weights. Variation in the texture pattern is encoded with the center pixel as a pivot for linking the neighbor pixel’s intensity within the block. The change in consideration of center pixels allows the encoding of different types of texture pattern. In LBP code formation, the specific order of pixels, i.e., anti-clockwise, is used to preserve the pattern information. The change in the order of the pixels causes variation in the generated code for the given block. Thus the configuration of these three parameters determines the encoding of the local texture pattern. Juefei-Xu et al. [17] formulated these LBP operations using the convolution operation. They convolved images with eight 3x3 convolution filters and performed binarization operations
to obtain the same results as obtained by LBP coding. All eight convolutional filters can be expressed as a 2-sparse difference filter. These filters are shown in Fig. (2). In standard LBP, all bits are weighted by the power of two and then accumulated to get the code of the respective block. Therefore, the convolution formulation of LBP can be expressed mathematically as,
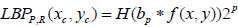 |
(2) |
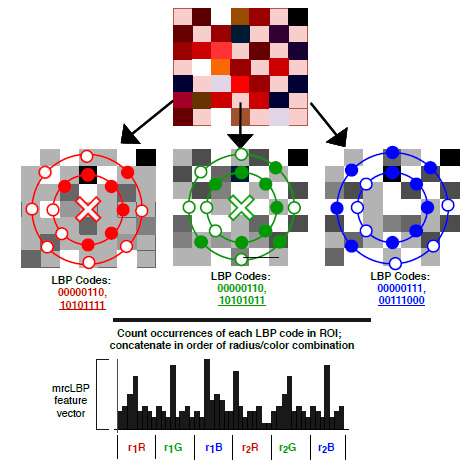
Where f (x, y) represents the convolution filters as shown in Fig. (2) for 8 neighbors, and H is the Heaviside function for thresholding operation. This formulation is used as a basis of CNN architecture in the proposed implementation. In the deep CNN architecture, a vector of an image is convolved with a filter followed by a non-linear like Rectified Linear Unit (ReLU), sigmoid, etc. The same operation is also obtained in LBP directed CNN architecture where convolution filters are used, as expressed in Fig. (2), and the Heaviside function is used as a non-linear operator. Secondly, basic LBP is operated on grayscale images. To fully utilize the information present in each color channel, we proposed the fusion of LBP code over all the color channels as expressed earlier [44]. The formulated LBP operator uses three-color maps, and fusion of these feature maps is used at every layer of the CNN architecture, as shown in Fig. (3).
The main advantage of the convolution formulation of LBP is that filters used in operation have a size of 1x1. Let “l and k” represent input and output feature maps respectively. The size of the convolutional filter is “n*m”. If p is intermediate channels, the ratio of the required parameters for standard CNN and LBP formulated CNN can be expressed by equation 3 [17]. If input feature map p is set equal to intermediate LBP channel m, then ration reduces to n × m. Thus it reduces the burden of CNN by a factor of 9 for 3x3 filter size. For detailed analysis, the readers are requested to refer to the paper that discusses it [17].
 |
(3) |
The local binary pattern framework in CNN is shown in Fig. (4). Fig. (4) shows the formulated layer structure using LBP filter, i.e., non-trainable filter and 1x1 convolutional filter, i.e., trainable filter. The proposed algorithm is tested on the BreakHis dataset for binary classification in benign and malignant tissue. Section 4 presents the experimental analysis.
4. RESULTS AND DISCUSSION
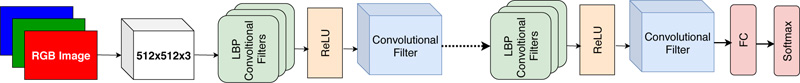
We configure the architecture of CNN to use LBP features and compare the obtained results with state-of-the-art methods. We consider the dataset of BreaKHis containing 2480 benign images and 5429 malignant images. All images are captured at four magnification factors, i.e., 40x, 100x, 200x, and 400x. All images have an RGB color map with a 700 x 460 resolution. To avoid overfitting problems, training images have been increased by the augmentation process using rotation, shifting, and mirroring. This augmentation process has been adopted in many CNN algorithms [45]. The model is based on the ResNet network. The filter size is fixed to 3x3 to incorporate LBP features formation in the first layer. The ReLU activation function is used after the first layer providing an LBP map and offering faster convergence in comparison with the Sigmoid function. The second layer contains trainable parameters with a size of 1x1 providing the feature map. The conceptual architecture of LBP directed CNN is shown in Fig. (5). Total 20 local binary pattern directed convolutional layers with filters of size 3 x 3 were used in the experiment. The regular CNN architecture with 20 layers requires approximately ~10.52 million learnable parameters, whereas LBP directed CNN architecture requires ~1.56 million parameters. Thus, the use of fixed LBP filters in CNN architecture reduces the parameters by 6.72 factor.
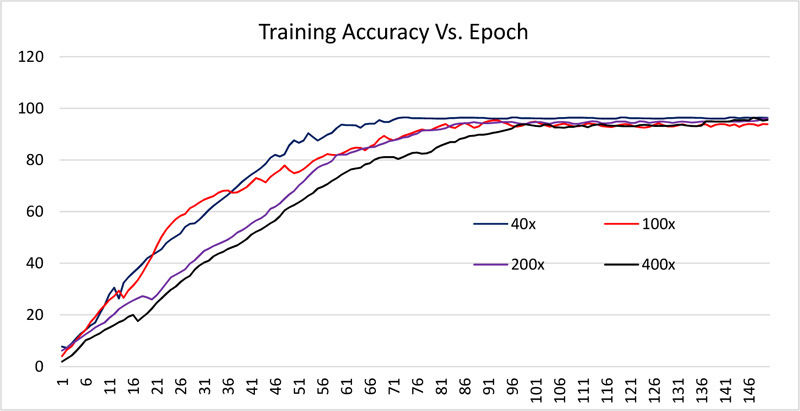
The dataset is divided into 70% and 30% for training and testing the network. All images are resized to 512 x 512. The network is trained using 150 epochs with 20 batch size. Received training accuracy and the testing accuracy is shown in Table 1 and Figs. (6 and 8). Table 1 demonstrates that the algorithm achieves higher accuracy for the 200x magnification factor in comparison with other magnification factors. Eventually, the maximum testing accuracy achieved with 200x magnification is 96.92% in comparison with training accuracy, i.e., 96.46%. From Figs. (6 and 8), it has been observed that the increment in the training and test accuracy is moderate after the 75 epochs. The receiver operating characteristic (ROC) gives the performance of the network at the various thresholds in terms of true positive rate versus false-positive rate. The area under the ROC graph (AUC) provides comprehensive performance measurement. The proposed network obtained more than 98% AUC. Further accuracy and loss analysis for both training and testing images are presented in Figs. (7 and 9).
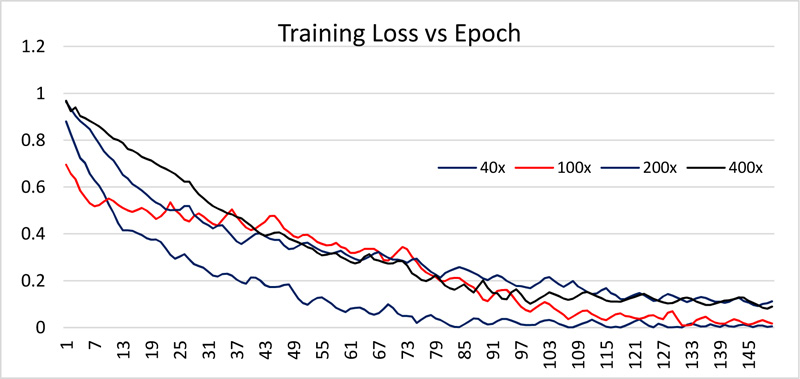
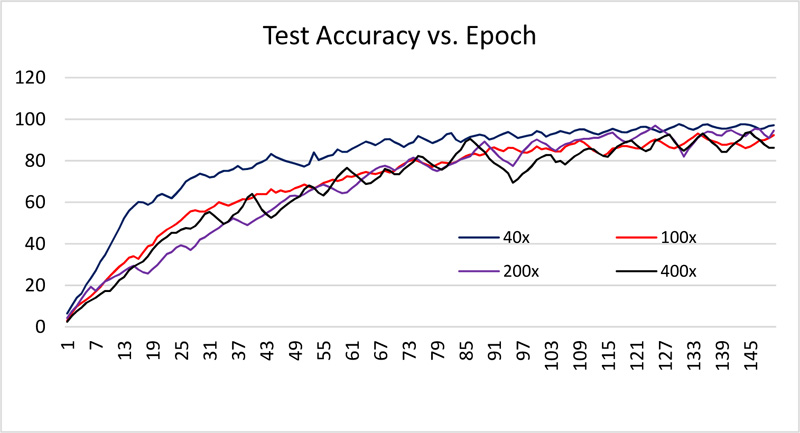
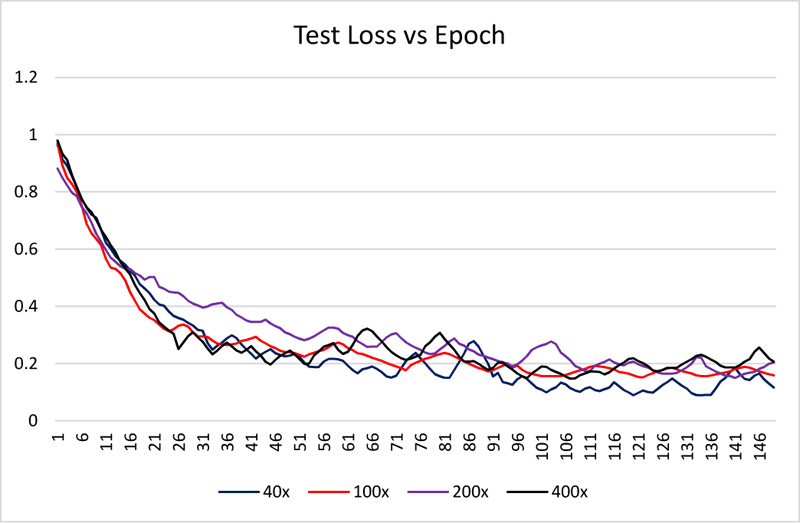
As classification accuracy evaluation does not provide the complete information, balanced information between the precision and recall, a measurement of classifier exactness and classifier completeness are equally important. Therefore, F1-Score giving a balance between precision and recall is demonstrated in Table 1.
|
Magnification Factor |
Accuracy (%) |
AUC (%) |
F1-Score (%) |
|---|---|---|---|
| 40x | 96.01 | 98.31 | 97 |
| 100x | 95.40 | 98.05 | 96 |
| 200x | 96.46 | 98.51 | 97 |
| 400x | 96.37 | 98.42 | 97 |
The combination of machine learning algorithms with features of LBP is not common in literature. Table 2 summarizes the finding on the BreaKHis dataset of histopathological images. Basic architecture like LeNET and ALEXNET architecture was able to succeed with 72% and 95.70% accuracy, respectively. The performance of the class structure-based deep convolutional neural network (CSDCNN) [46] is better than traditional networks with an average accuracy of 93.25% in the presence of a large number of parameters due to deep structure. In another research [34], the author tested VGG16 and ALEXNET network with LBP features. The feature length is quite high, i.e., 4096. VGG16 was speciously superior than ALEXNET with an accuracy of 94.72%. In their proposed approach, LBP features were classified using histogram intersection kernel-based SVM (IKSVM). All images are resized to 512 x 512 in the process, and obtained accuracy is 96.50. The direct comparison cannot be established because they have used the SVM approach and the dataset is also different. However, the proposed algorithm’s accuracy is similar with a reduction in the training parameters by the factor of 9. Bayesian network [31] has been tested on MIAS datasets consisting of gray-scale mammogram images with accuracy limited to 92%. Spanhol et al. [47] also proposed a CNN network with maximum accuracy of 90% achieved on the dataset with a 40x magnification factor. However, for another magnification factor, the accuracy level is limited to 88.40% only, limiting the average accuracy to 87.27%. The fusion of VGGNET and Random Forest (RF) was also used [48] for binary classification of BreaKHIS dataset and achieved 93.41% accuracy. A multi-task exception CNN was presented with fine tuning of the hyperparameters [49]. The performance of their network is better than the single task CNN with 93.36% of accuracy. The ALEXNET CNN architecture has 2.33 million learnable parameters [17, 50] and VGGNET16 architecture has 15 million learnable parameters. The proposed network achieved 96.46% on images with a 200x magnification factor with 1.5 million learnable parameters. Table 3 summarizes the required learnable parameters by each model.
CONCLUSION
In this paper, we demonstrated the use of LBP features in the CNN network. The formulation of LBP using eight convolution filters makes the model simpler with fewer learning parameters. The study proposes that texture features play a significant role in the histopathological image classification and LBP generates the most robust rotation and illumination invariant features in comparison with other texture features operators. Recent deep CNN architectures have achieved comparable classification accuracy at the cost of high training parameters or computational complexity. Therefore, the suggested alternates reduced the burden of CNN architecture and preserved the accuracy level. This makes it suitable for the environment having fewer resources. The proposed algorithm obtained an average accuracy of 96.06% on the BreaKHis dataset. This validated the proposed network’s performance. The performance achieved can be further tested on other datasets as well as mammogram images.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The author declares no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.


