All published articles of this journal are available on ScienceDirect.
Isolation and Characterization of an Aeromonas punctata Bacteriophage
Abstract
An Aeromonas punctata bacteriophage, named as DH1, was isolated from East Lake, Wuhan city, China. Morphologically, phage DH1 showed a typical Myoviridae structure consisting of an isometric head (50 nm in diameter) and a visible tail. The bacteriophage had a latent period of about 90 minutes and an average burst size of about 125 PFU•Cell-1. Restriction enzyme pattern of the bacteriophage’s genome showed that the genome is a double-stranded DNA and about 34kb in size. The sequenced genomic fragments showed highly similarities to gp04 and gp16 sequence of other Myoviridae bacteriophages at protein level.
1. INTRODUCTION
Aeromonas sp. has been well known to cause diseases in fishes, frogs, and even humans [1-4]. Till now, only one Aeromonas hydrophila bacteriophage has been isolated [5] and sequenced [6]. In this study, we reported the isolation and characterization of a novel Aeromonas punctata bacteriphage from a natural lake.
2. MATERIALS AND METHODS
Sampling: The sampling site was located in East Lake (30˚31′37.66″N,114˚22′09.18″E), Wuhan City, Hubei Province, China. Surface water samples (10-20cm depth) were collected and transported to laboratory immediately for bacteria and virus isolation during December 23, 2006 to January 8, 2007,
2.1. Isolation of Bacteria and Bacteriophage
A volume of 100μL water from lake was spread over solid LB media. After incubation at 37℃ for 24h, separated colony was picked and purified by standard methods. The isolation of bacteriophage was performed using phage plaque method. Briefly, 30mL fresh lake water was filtered through a 0.2μm filter (PN4612,PALL). A volume of 100μL filtered water was mixed with 200μL bacterial culture in LB media and incubated at 28℃ till the plaque was formed. Pure bacteriophage strain was obtained by five serial single-plaque isolations. Only one phage-host system was established from 60 purified bacterial colonies, the phage was named as DH1, and the host bacterium was designated MH1.
2.2. Characterization of MH1
The genomic DNA of bacteria MH1 was extracted with phenol-chloroform-isoamylol and precipitated with ethanol. A fragment of 16S rDNA was amplified by one pair of universal primer [7] and sequenced.
2.3. Phage Purification
Five hundred milliliters of cell lysates (the titer of the phage was about 109PFU•mL-1) was centrifuged at 10000 g for 30 min in Jouan KR2.5i centrifuge to remove the cell debris. Then, the supernatant was centrifuged at 100,000 g for 2.5 h in Beckman J-30I (JS-24 rotor) to concentrate the phage stock to about 2mL. The phage stock was further purified on 20%-50% sucrose density-gradient centrifugation by Beckman optimaTm MAX and MAX-E(MLS-50rotor). Finally, the phage stock was diluted in 3mL 20mmol/L MgCl2 solution and stored at 4°C.
2.4. Transmission Electron Microscopy (TEM)
The purified phages were dropped onto a formvar carbon-coated copper grid (200 mesh) and negatively stained with 2% sodium phosphotungstate (pH7.0) and observed under TEM (Hitachi H-7000FA).
2.5. Phage DNA Extraction and Restriction Endonuclease Digestion
The nucleic acid of purified phage was extracted using protease K, followed by phenol-chloroform-isoamylol extraction and ethanol precipitation. The phage genomic DNA was digested with endonuclease AccⅠ, BamHⅠand Sau3AⅠ (Takara) and load onto a 0.8% agarose gel and analyzed using Ethidium Bromide staining.
2.6. Cloning and Sequencing
Two BamHⅠfragments, 1.4 kbp and 1.9 kbp were purified with a gel extraction kit (D2501-01, OMEGA), then cloned to pUC19 vector using standard protocol [8]. The recombined plasmids DNA were purified and sequenced with universal primers by Invitrogen Company.
3. RESULTS AND DISCUSSION
3.1. Identification of the Host
A fragment of 196 bp of 16S rDNA was amplified and sequenced from bacteria MH1, and the sequence showed 100% identity to Aeromonas punctata and was submitted to GenBank (access number: EU515214).
3.2. Identification of the Phage
TEM analysis showed that phage DH1 had an icosahedral head (50 nm in diameter) and a visible tail (less than 100 nm in length) (Fig. 1), which is morphologically similar to but much smaller than Aeromonas hydrophila phages Aehl and Aeh2 [5].
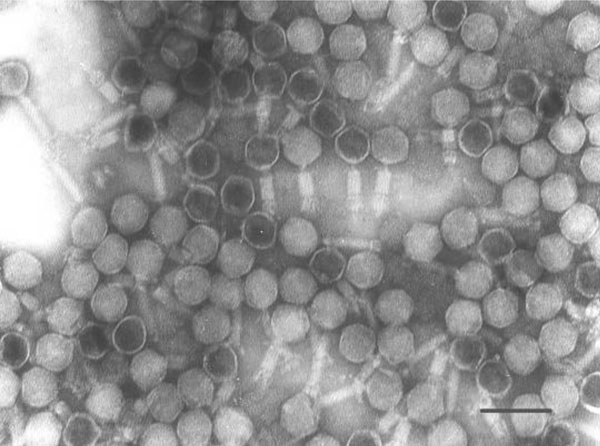
Electron micrograph of negatively stained phage DH1. Bar: 100nm
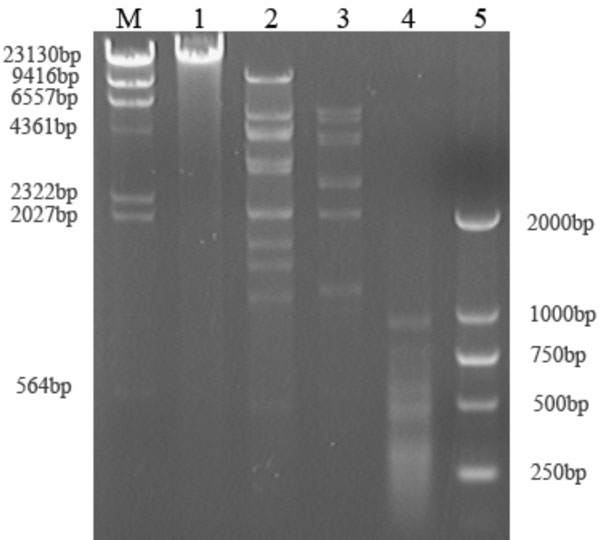
Restriction enzyme digested patterns of DH1 genome DNA in 0.8% agarose gel electrophoresis.
M: λ phage DNA digested by HindIII, 1: phage genome DNA without digestion, 2: Acc I, 3: BamH I, 4: Sau3A I, 5:DL2000 marker.
3.3. Genome Analysis of DH1
The result of enzyme digestion showed that the genome size is approximately 34±3kb (Fig. 2) and much lesser than Aeh1(230kb)[9]. The sequence of two fragments, 1.4 and 1.9kb, were submitted to GenBank (access number: EU515215 and EU515216) and showed 42% and 50% identities to gp04 and gp16 of a Myoviridae bacteriophage Becp43, respectively [10].
3.4. Growth Cycle
The one-step growth curves of DH1 indicated that the latent periods of phage DH1 were 90 min, which was much longer than Aeromonas hydrophila phages Aehl and Aeh2 [5]. The average burst sizes of phage DH1 were about 125 PFU•Cell-1, which was also bigger than Aehl and Aeh2 [5].
The majority of Aeromonas sp. described to date are from soil and water and known to be pathogenic to cold-blooded animals. Some studies have demonstrated that the presence of A. sp. in drinking water is a potential risk, since some strains can produce a wide range of virulence factors [2, 4]. Thus, by studying the interaction of the phage-host system, A. sp. phage may be significant for controlling the risk [11].
CONCLUSION
A. punctata bacteriophage DH1 was a typical Myoviridae bacteriophage, and its genome is a 34kb double-stranded DNA. The sequenced genomic fragments showed highly similarities to other Myoviridae bacteriophages.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
This work was supported by National Science Foundation of China [grant numbers 31200385,> 31370148] and Science Foundation of Hubei Province [2013CFA108].


