All published articles of this journal are available on ScienceDirect.
Anatomically Based Geometric Modelling Using Medical Image Data: Methods and Programs
Abstract
Geometric modeling software that can realize two-dimensional medical image browsing, preprocessing, and three-dimensional (3D) reconstruction is designed for modeling human organs. This software performs medical image segmentation using a method that combines the region growing and the interactive segmentation methods. The Marching Cubes surface reconstruction algorithm is used to obtain a 3D geometric model. The program is compiled using Visual Studio 2010. The software is employed to obtain the geometric model of the human femur, hipbone, and muscle. The geometric modeling results can accurately express the structural information of the skeleton and muscle.
1. INTRODUCTION
Our research team has been working on the virtual surgery system. The three-dimensional (3D) geometric modeling of the human tissue or organ is the core module of the virtual surgery system. The 3D geometric model can aid doctors in diagnosing illnesses accurately and efficiently, which helps in improving the success rate of surgery. In this work, the methods used to design the geometric modeling software in the virtual surgery system have been introduced. The 3D geometric modeling of the human tissue consists of the preprocessing and 3D reconstruction of medical images. Herein, the core operation in the image preprocessing is the segmentation of medical images. R. Kayalvizh et al. proposed a new intelligent algorithm referred to as particle swarm optimization [1]. I. Cruz-Aceves et al. proposed an automatic image segmentation method based on an active contour model theory and estimation of distribution algorithms [2]. N. R. Nayak et al. proposed an improved clustering algorithm, which is applicable to gray images, by evaluating three different kinds of clustering algorithms [3]. For general medical images, it is difficult to achieve the required accuracy using automatic segmentation methods. Therefore, an interactive operation is required in image segmentation to obtain results that are more accurate. M. Milanova et al. proposed an image segmentation method based on vision to determine location, which applied the Chan-Vess model to the image segmentation [4]. Z. T. Liu et al. proposed a novel method for complex medical image segmentation based on the hierarchical characteristics of human visual perception [5]. R. Delgado-Gonzalo et al. proposed an interactive image segmentation method based on the B-spline curve [6]. A. Chavez-Aragon et al. proposed a network platform that collected the MRI images of volunteers to form a knowledge-based image segmentation of the hip [7]. Although many methods of image segmentation have been proposed in recent years, it is difficult to obtain satisfactory results using any individual method of the computer image segmentation, whether automatic or interactive, because of the limitations and difficulties in segmentation method. In this study, we combined the region growing method, which is based on the gray level difference, and the interactive segmentation method, which is based on the edge fill algorithm, the combination of the region growing method and interactive segmentation satisfied the general requirements of medical image segmentation.
Many 3D reconstruction methods have been proposed in the past [8-11]. The MC (Marching Cubes) algorithm has been used in this study. Compared to other algorithms, the MC algorithm is easily programmable and generates 3D models at a fast speed. Besides, the method, which utilizes ordinary computer graphics hardware to implement rapid geometric transformation and mapping, occupies less memory and has high portability. Furthermore, the surface structure of the human tissue can be expressed accurately to meet the requirements of the virtual surgery system.
Based on the region growing module, the interactive segmentation module, and the 3D reconstruction module, the geometric modeling software was created for human tissues or organs, which included two-dimensional (2D) medical image browsing, preprocessing, and 3D geometric reconstruction. This software can be used both independently and as a part of the virtual surgery system. The features of the software are as follows: (1) Medical images can be imported and then panned and zoomed for convenient observation. (2) Different medical image segmentation methods can be used. Image enhancement, noise reduction, segmentation, erasing and patching vulnerabilities for medical images can be conducted using the software. (3) 3D reconstruction can be carried out after preprocessing the images. Then, the 3D geometrical model can be displayed on the interface. The model can be zoomed, rotated, moved, etc. (4) The Smooth handling of the surface of the 3D geometrical model can be conducted easily to make it closer to real human tissues and organs. (5) The cutting of an arbitrary cross-section can be performed in the 3D geometrical model for the users to observe. (6) Data files can be obtained in multiple formats and exported to other software.
2. METHODS
C++ is employed to design the geometric modeling interface. The core modules of the software include the region growing module, the interactive segmentation module, and the 3D reconstruction module.
2.1. The Region Growing Module
The region growing method, in which the gray value is used as a similarity measurement criterion, is utilized to segment 2D medical computed tomography (CT) images in this module. The human tissues can be segmented based on the parameters of the growth criterion. This module is applicable for segmenting medical images for which the gray value is uniform, the difference in gray value between tissues is apparent, and the edge is distinct. For the tissues whose gray value difference is small or the distribution of the gray level is non-uniform, cavity and excessive segmentation form easily. Therefore, the interactive segmentation module can be combined with the region growing module to perform segmentation.
The basic idea of the region growing algorithm is that pixels with the same or similar nature are gathered to constitute a region. First, in the region of interest, a pixel point is artificially selected as a seed point. Then, based on the growth criterion, the pixels that meet this criterion are incorporated around the seed points into a seed region. Then, these new pixel points are taken as the seed points and the abovementioned steps are repeated until there are no pixels left that meet the growth criterion. In this manner, the region can be formed [12, 13].
The key to the region growing algorithm is the growth criterion and the selection of seed points, which is not only related to a specific segmentation problem, but also to the type of medical data to be segmented. The growth criteria may be based on different principles. The difference in the growth criteria affects the process of region growing and the segmentation results. Most of the growth criteria are based on the local characteristics of images. In this study, 2D medical images have been used. Consequently, the selected growth criterion is based on the regional gray level difference. The flow of this algorithm is as follows: A typical point in a region of the image is selected as the seed point based on the purpose of segmentation. After initializing the stack of computer memory, the specified seed point is stored into the stack. Then, a member is chosen from the stack top and eight of its neighborhoods are scanned. When the gray value of the pixel satisfies the equations,
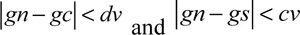
(gs is gray level of seed point. gn is gray level of neighbor point. gc is gray level of current neighborhood pixel point), the value is stored into the stack and the cycle is repeated until there are no pixels left that meet the growth criterion.
2.2. The Interactive Segmentation Module
The function of this module is to segment the loaded 2D CT images using the interactive segmentation method. A polygon is drawn manually. Then, the region of interest is selected, extracted, and separated to perform the segmentation of medical images. Segmentation of regularly shaped tissues or organs is better and takes comparatively less time. For segmentation of irregularly shaped tissues, the region growing module is frequently employed along with the interactive segmentation module. Otherwise, the segmentation task takes more time.
In this study, the interactive segmentation method is based on the edge fill algorithm. Based on the segmentation region, an operator can plot an arbitrary polygon. Then, the area surrounded by the polygon can be automatically separated from the whole region using the algorithm. The flow of interactive segmentation is as follows: the images are initialized, i.e., all the points used in the maker image are initialized to zero. Then, each edge is discretized, i.e., the direction of the scan line is set as the horizontal direction. The separation distance between two adjacent scan lines is seen as the unit distance of the image in the vertical direction. A non-zero value is assigned to the intersections of the polygon and each scan line. Then, the polygon is filled. All the points that are on the right side of each non-zero point are taken as complements. Then, the segmentation result is generated. For non-zero points in the maker image, the pixel points in the exported image are equal to the value of the corresponding points in the imported image. The corresponding points in exported image are also assigned as zero, for zero points in the maker image.
2.3. The 3D Reconstruction Module
The classic MC algorithm is employed as the surface reconstruction algorithm. Users can isolate the tissues between two thresholds by setting a high threshold and a low threshold to implement the surface reconstruction of medical images. This module makes use of the 3D reconstruction algorithm based on voxels to carry out the 3D reconstruction for a set of 2D medical CT images after segmentation. In addition, users can zoom, move, and rotate the 3D model to conveniently observe the tissue structure, the size of the focus, the location, and other information about the tissue. The flow diagram of the 3D reconstruction algorithm is shown in Fig. (1).
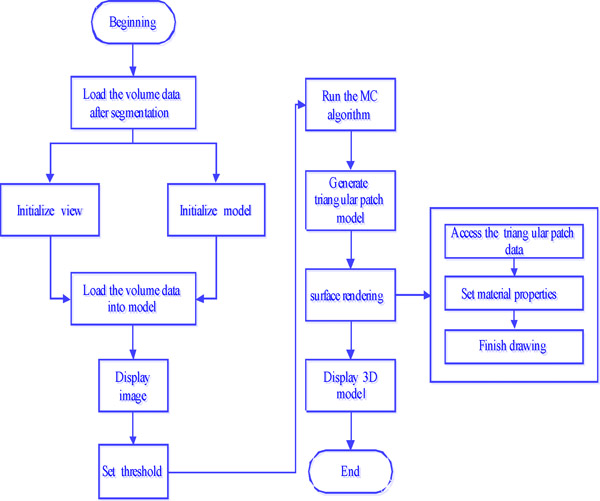
3D reconstruction program flow chart.
3. RESULTS
The medical image data is derived from the CT images of the hip of a healthy man, whose age is 26 years, height is 175 cm, and weight is 65 kg. He is in good condition and has no history of hip fracture, muscle damage, etc. Axial continuous tomography is used in this work. Seven hundred tomographic images are selected. The space between the images is 1 mm. Every image is a lattice of 512×512 pixels. In addition, Visual C++ is used in the geometric modeling program, which is run on the Windows platform.
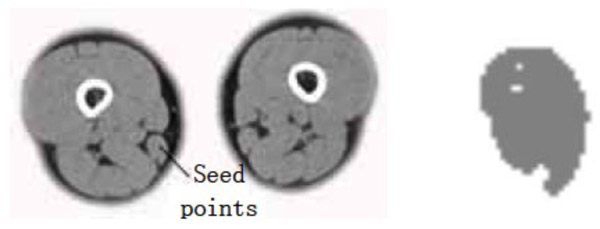
Gracilis muscle segmentation result (dv=cv=30).
The program results include the following: (1) The segmentation results of the object under different parameters and the same seed points, and under the same parameters and different seeds points obtained using the region growing algorithm. (2) The segmentation results of the femur and the semitendinosus obtained using the region growing algorithm and the interactive segmentation algorithm.
- (1) The points selected in the source gracilis image are taken as the seed points. The value of dv and cv is taken as 30. The segmentation result can be obtained using the region growing method as shown in Fig. (2).
- (2) The points selected in the source gracilis image are taken as the seed points. The value of dv and cv is taken as 90. The segmentation result can be obtained using the region growing method as shown in Fig. (3).
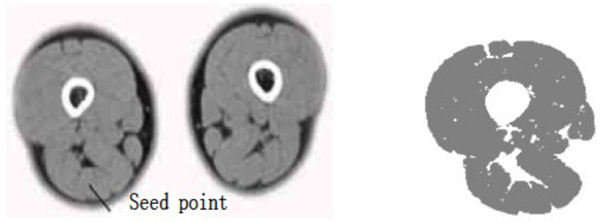
- (3) The points selected in the source semitendinosus image are taken as the seed points. The value of dv and cv is taken as 30. The segmentation result can be obtained using the region growing method as shown in Fig. (4).
- (4) A point selected in the source femur image is taken as the seed point. Owing to the larger gray value of the bone, the value of dv is 300 and that of cv is 900. The segmentation result is shown in Fig. (5).
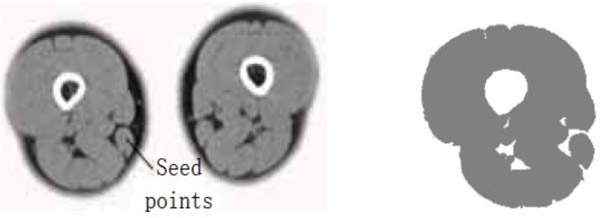
Gracilis segmentation result (dv=cv=90).
Semitendinosus segmentation result (dv=cv=30).
Femur segmentation result.
The segmentation result shows that for the case where the points selected in the source gracilis muscle image are taken as the seed points, and the value of dv and cv is 30, the whole gracilis muscle section can be obtained using the region growing algorithm. However, for the same seed points, when the value of dv and cv is 90, owing to this larger value, multiple connected muscles are obtained using the region growing method. If the value of dv and cv is 30, then users can select a point in the semitendinosus as the seed point for segmentation. The segmentation result shows multiple muscles including the semitendinosus because the boundaries of the semitendinosus are fuzzy. As observed from the segmentation result of the muscle and the bone, the region growing algorithm put forward greater demand in the control of parameters and the selection of seed points. In addition, for the tissues whose boundaries are fuzzy, it is easy to extract excess parts along with the segmentation object, which results in over-segmentation.
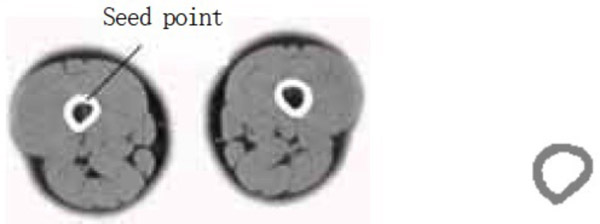
- (5) The sources of medical data are the same for the region growing module, i.e., users utilize the interactive segmentation algorithm to conduct the segmentation of the muscle and the bone. The segmentation results of the semitendinosus and the femur are shown in Figs. (6 and 7), respectively.

Muscle segmentation result.
Femur segmentation result.
As observed in the segmentation result for case (5), users can take advantage of the interactive segmentation method to segment any tissue and organ of interest. However, the method has a high operating time. Furthermore, it places greater demand on the operator’s skill. Therefore, the tissues whose boundaries are clear and the gray level difference is high, such as the femur, can be segmented immediately using the region growing method. Comparatively, the muscle tissues whose boundaries are fuzzy, such as the semitendinosus, the interactive segmentation method can be used. Users can also employ the interactive segmentation method first to obtain the parts of interest. That is, the tissues whose gray level difference is small are separated. Then, users can employ the region growing method to obtain the desired results. This not only avoids the over-segmentation that results from the region growing method, but also improves the efficiency of the segmentation.
4. DISCUSSION
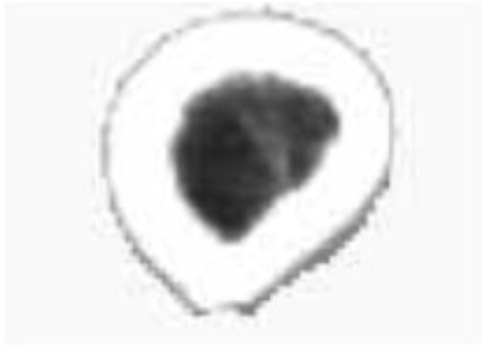
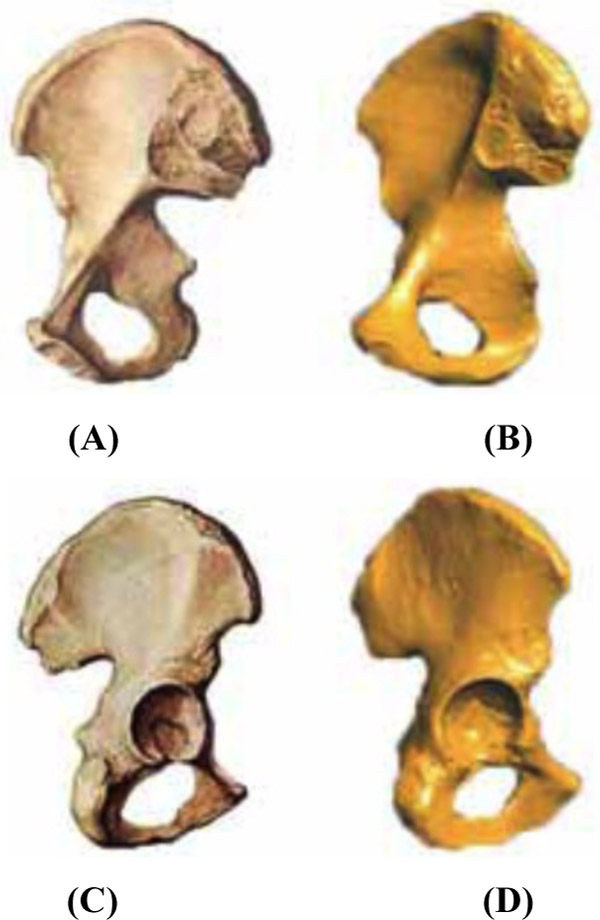
Since the 3D reconstruction model is created using a computer, which gives an unnatural image, users have to set and adjust the relevant parameters as needed. As a result, it is difficult to assess the results of the reconstruction quantitatively. At present, the assessment of the quality of the 3D reconstruction is mainly subjective. Based on the experience in anatomy or comparison with real human organs, users compare key feature points of the 3D model to judge the merits of the reconstruction results. The comparison between the 3D model reconstructed by our software and the structure of the real muscle and bone is shown in Figs. (8 and 9).

Comparison of human sartorius muscle and muscle geometry model.
Comparison of human hip and hip geometry model.
The comparison for the sartorius muscle is shown in Fig. (8). The real structure of the human sartorius and the geometry model of the sartorius muscle obtained using the software are shown on the left and right, respectively.
The comparison for the hipbone is shown in Fig. (9, 9(a) shows the front structure of the real hipbone. Fig. 9(c) shows the back of the real hipbone. Fig. 9(b, d) show the model for the front and back of the hipbone, respectively, obtained using the software.
As observed from Figs. (8 and 9), the 3D geometry model obtained using this software joins basically with the physiological structure of real muscle and bone and can show up the detailed characteristics of corresponding muscle and bone. In future studies, the generation of a volume model and the implementation of the program will be considered for integration with the meshing module and the finite element analysis of the biomechanics module that our research group has designed.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
This research was supported by NSFC (No. 61572159), NCET (NCET-13-0756), Scientific Research Foundation for the Returned Overseas Scholars of Heilongjiang Province and Distinguished Young Scientists Funds of Heilongjiang Province (JC201302).


