RESEARCH ARTICLE
Association Between KCNJ11 Gene E23K Polymorphism and Body Composition Together with Its Response to Endurance Training
Zhou Duoqi1, *, He Qing1, Hu Yang2, Li Yanchun2, Xi Yi3, Wen Li3
Article Information
Identifiers and Pagination:
Year: 2015Volume: 9
First Page: 121
Last Page: 125
Publisher ID: TOBEJ-9-121
DOI: 10.2174/1874120701509010121
Article History:
Received Date: 3/12/2014Revision Received Date: 20/2/2015
Acceptance Date: 2/3/2015
Electronic publication date: 29/5/2015
Collection year: 2015

open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
Abstract
To explore the association between KCNJ11 gene E23K polymorphism of Chinese and body composition together with its response to endurance training. 102 biologically unrelated Han nationality male new recruits from northern China volunteered to execute a 5000-m running program, and the intensity is 95–105% individual lactate threshold. The protocol was lasted for 18 weeks, three times per week. The body composition index, including body weight (WT), lean body weight (LBW), body mass index (BMI) and body fat percentage (Fat%), was measured before and after training. PCR-RFLP was used to detect the KCNJ11 gene E23K polymorphism. Hardy-Weinberg equilibrium was observed for the frequency of genotypes in these subjects. Before training, WT, BMI and Fat% in KK group were significantly higher than those in EE and KK group (P<0.05 or P<0.01). There was no significant difference in LBW among groups (P>0.05). After training, the changes of all body composition index in KK group were bigger significantly greater than those in EE and EK groups (P<0.01). KCNJ11 gene E23K polymorphism might contribute to individual body composition together with its response to endurance training. The body fat content at baseline in KK was more than those in EE and EK groups, and it may hinder that individual to eliminate their body fat during endurance training.
1. INTRODUCTION
The human ATP-sensitive potassium (KATP) protein-coding genes KCNJ11 (or Kir6.2) subunit is located on chromosome 11P15.1, having a total length of 3418 bp, containing only a 1173 bp exon, which is widely expressed in heart, skeletal muscle, vascular smooth muscle, nerve, and various endocrine tissues in human [1-3]. KATP couples cellular energy metabolism and bioelectrical activity, and thus participates in important functions such as in energy metabolism, regulate hormone secretion, sports fatigue and cell protection [4-6]. The study found that KCNJ11 gene E23K polymorphism can inhibit insulin secretion, thereby increasing the secretion of glucagon, in diabetes and elevated glucagon related disease [7, 8]. Insulin and glucagon play important roles in glucose and lipid metabolism in vivo. Therefore, it is now presumed that KCNJ11 gene E23K polymorphism may be associated with the body composition.
Endurance training can improve insulin sensitivity, and speed up the body's glucose and lipid metabolism; furthermore, the study found that individual difference existed in the effect of endurance training, which is influenced by genetics. Therefore, the purpose of this study is to investigate the relationship between body composition sensitivity to endurance training, and KCNJ11 gene E23K polymorphism by training 102 new recruits, who execute a 5000-m running program for 18 weeks. This study would provide a molecular target for further research and development of personalized exercise prescription.
2. SUBJECTS AND METHODS
2.1. Subjects
The volunteers were 102 biologically unrelated healthy males newly recruited from northern China, without the experience of endurance training. The age was 18.8 ± 0.91 years, the height was 171.8 ± 5.9 cm. Written informed consent was obtained from each participant.
2.2. Methods
2.2.1. Training Protocol
All participants performed a 5000 m running program. Heart rate sporttesters (S810, Finland) were used to monitor constant training heart rates corresponding to individual lactate threshold. During 1–10 weeks, the volunteers were trained with an intensity of 95% individual lactate threshold. Then they were trained with an intensity of 105% individual lactate threshold during the last 10 weeks. The adaptation training for 2 weeks was completed before formal training. The participants performed this program three times per week lasting for 18 weeks without other, endurance training.
| Genotype | P value | |||
|---|---|---|---|---|
| EE | EK | KK | ||
| n | 35 | 54 | 13 | |
| before H(cm) |
170.23±6.32 |
172.15±5.40 |
173.54±5.68 |
NS |
| WT(kg) | 58.91±4.20 | 60.65±3.78 | 65.51±4.22 | 0.000 |
| LBW(kg) | 36.68±3.11 | 38.09±3.61 | 36.78±3.08 | NS |
| BMI | 20.34±1.24 | 20.48±1.27 | 21.76±1.04 | 0.002 |
| Fat% | 14.46±1.77 | 14.48±1.79 | 15.73±0.88 | 0.050 |
| after H(cm) |
170.69±6.22 |
172.39±5.23 |
174.23±5.21 |
0.000 |
| WT(kg) | 59.52±4.35 | 61.35±3.67 | 67.89±4.12 | 0.000 |
| LBW(kg) | 36.91±3.35 | 36.90±3.57 | 37.58±3.05 | 0.000 |
| BMI | 20.41±1.18 | 20.64±1.18 | 22.37±1.07 | 0.000 |
| Fat% | 15.11±1.97 | 15.46±1.81 | 18.37±1.33 | 0.000 |
2.2.2. Genotype Analysis
DNA was extracted from lymphoblastoid cell lines by Wizard® Genomic DNA Purification Kit (Promega, USA). Primer pairs for PCR-RFLP of the KCNJ11 gene E23K polymorphism were designed by Primer 5.0 software:
Forward: 5’-gac tct gca gtg agg ccc ta- 3’;
Reverse: 5’-acg ttg cag ttg cct ttc tt- 3’;
The amplification protocol was according to Hansen [9]. Preventive contamination measures were taken by the inclusion of PCR reaction mixture without DNA (negative control). Final amplification product was digested by 8U restriction enzyme Ban II (Takara, Japan) for 3h at 37°C. The resulting fragments were separated by horizontal electrophoresis on 4% agarose gels at 100V for 40 minutes, and photographed under UV transmitted lights.
2.2.3. Body Composition Index Test
All body composition index, including body weight, lean body weight, body mass index and body fat percentage, was measured by Tanita-DX200 (Japan) before and after training.
2.2.4. Statistical
Pearson Chi-square was used to determine whether the observed genotype frequencies were in Hardy-Weinberg equilibrium. Differences in body composition index between three genotype groups were detected by ANOVA's Post Hoc test. Analysis of covariance was used to test the differences of changes in body composition index after training between three genotype groups P<0.05 which were considered statistically significant. All statistical analyses were performed with SPSS software for Windows 11.5 package.
3. RESULTS
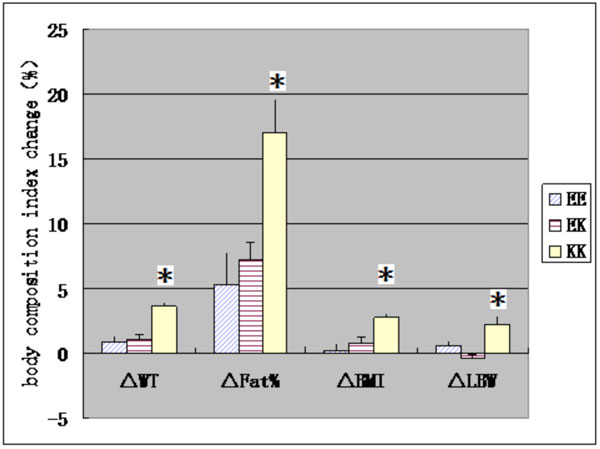
Asterisks mean significant difference in body composition index before training or changes of body composition index after training between three genotype groups.
After PCR and digested by Ban II restriction enzyme, three genotypes EE (35, 34.3%), EK (54, 52.9%) and KK (13, 12.8%) were obtained. The frequency of genotypes was in Hardy-Weinberg equilibrium (χ2 = 1.245, P> 0.05).
Table 1 shows that except LBW, all body composition index in baseline before endurance training including WT、BMI and FAT% in KK group was significantly higher than those in EK and EE groups (P <0.05 or P <0.01). No significant difference was observed in LBW in baseline before training between three genotype groups (P>0.05).
After training, all body composition indexes except LBW increased significantly. There are significant differences of changes in body composition index after training between three genotype groups (P <0.01, Table 1). Furthermore, covariance analysis found that the changes in all body composition index after training in KK group were significantly greater than those in EE and EK groups (P <0.01, Fig. 1).
4. ANALYSIS AND DISCUSSION
4.1. Association Analysis of Body Composition, KCNJ11 Gene E23K Polymorphism
This is the first study to explore the relationship of body composition and its sensitivity to endurance training and KCNJ11 gene E23K polymorphism of Han population from plains of northern China. The results showed that except LBW, all body composition index in baseline before endurance training including WT, BMI and FAT% in KK group, was significantly higher than those in EK and EE groups. Considering that no significant difference was observed in LBW in baseline before training between three genotype groups, we speculated that differences in WT and BMI maybe due to the differences in FAT%. Therefore, since we believe KCNJ11 gene E23K polymorphism are related to body fat content, subjects carrying KK genotype may have a higher body fat content. Li L’s studies have shown that K allele can regulate the PH value in skeletal muscle and increase the chance of KATP opening, leading to the cell hyperpolarization, decreased cell excitability and reduced skeletal muscle glucose uptake and use; excessive sugar in the body is easily converted into fat [10]. Meanwhile, K allele because of reduced cell excitability, also decreases skeletal muscle contraction and energy consumption, leading to the energy surplus which contributes to fat synthesis [11]. Kanezaki Y. observed the significant weight gain and obesity in kir6.2 knockout mice because of insulin resistance in skeletal muscle and adipose tissue [12].
The possible mechanism of KCNJ11 gene E23K polymorphism effects body composition and may arise from E mapped in the loop region of the N-terminal IC domain, proceeded by the first helix and the critical slide helix. Whether the channel protein is open is determined by their interaction. Negatively charged E23 positioned planar to these two Helices. The K23 variant would not be directly involved in structural distortion. K23 may disrupt the integrated interaction because of unbalancing charges [13, 14] (Fig. 2). Previous researches in vitro showed that E23K polymorphism is related to the regulation sensitivity of KATP channel protein to ATP, inhibits insulin secretion [15, 16], reduces sugar intake and utilization, and accelerates the accumulation of fat.
4.2. Association Analysis of the Effects of Endurance Training on Body Composition and KCNJ11 Gene E23K Polymorphism
Our study found that WT, BMI and Fat% after endurance training were significantly increased. This is different from some previous results that showed that the endurance training can reduce fat. The reason may be that the subjects are energetic in late puberty (average age 18.8 years old). The endurance training load (5000 m training for three times a week) in this study was not sufficient to stimulate the body to break down a lot of fat for energy, and while exercise increases the appetite, excessive intake of calories, resulted in the excessive energy that will be stored for fat synthesis. Moreover, development, nutrition, and pace of life may contribute to this result [17]. Our study also found a significant increase in height of the subjects after training (P <0.05, Table 1).
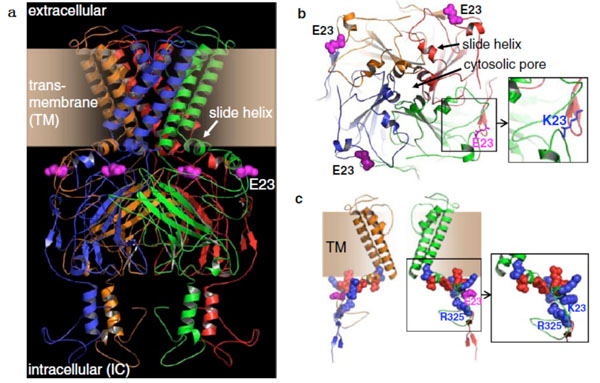 |
Fig. (2). Molecular model of Kir6.2 channel pore (cited by reference 13) a side face b above face c Structure of two Kir6.2 subunits. |
The study found that KCNJ11 gene E23K polymorphism may be associated with effects of endurance training on body composition, the changes of all body composition index after training in KK group were significantly greater than those in EE and EK groups. Considering that no significant change was observed in LBW after training, we speculated that differences of changes in WT and BMI after training between three genotype groups maybe due to those in FAT%. One reason for this difference may be the inhibition of insulin secretion [15, 16] through K allele. The other possible reason is that the endurance training can increase the proportion of slow-twitch fibers. Banas K found that the concentration of KATP channel protein in slow-twitch fibers was significantly lower than those in the fast-twitch muscle fibers [18]. Therefore, endurance training may reduce the KATP channel protein in slow-twitch fibers, thus decreasing fat expenditure. Kanezaki also confirmed that kir6.2 may be associated with obesity by knockout mice experiment [12].
CONCLUSION
KCNJ11 gene E23K polymorphism might contribute to individual body composition together with its response to endurance training. The body fat content at baseline in KK were more than those in EE and EK groups, and it may hinder the individuals to eliminate their body fat during endurance training.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
This work was supported by Department of Science and Technology of China (2003BA904B04) and Natural Science Foundation of Anhui province (1308085MH161).