All published articles of this journal are available on ScienceDirect.
Exploring Biomedical Named Entity Recognition via SciSpaCy and BioBERT Models
Abstract
Introduction
Biological Named Entity Recognition (BioNER) is a crucial preprocessing step for Bio-AI analysis.
Methods
Our paper explores the field of Biomedical Named Entity Recognition (BioNER) by closely analysing two advanced models, SciSpaCy and BioBERT. We have made two distinct contributions: Initially, we thoroughly train these models using a wide range of biological datasets, allowing for a methodical assessment of their performance in many areas. We offer detailed evaluations using important parameters like F1 scores and processing speed to provide precise insights into the effectiveness of BioNER activities.
Results
Furthermore, our study provides significant recommendations for choosing tools that are customised to meet unique BioNER needs, thereby enhancing the efficiency of Named Entity Recognition in the field of biomedical research. Our work focuses on tackling the complex challenges involved in BioNER and enhancing our understanding of model performance.
Conclusion
The goal of this research is to drive progress in this important field and enable more effective use of advanced data analysis tools for extracting valuable insights from biomedical literature.
1. INTRODUCTION
Named Entity Recognition (NER) [1] is a crucial part of Natural Language Processing (NLP) that involves identifying and categorising named items in written text. Named entities encompass several categories such as personal names, organisational names, and geographical places, among others. The primary objective is to do this by transforming unstructured textual content into a machine-readable format. Named Entity Recognition (NER) is a crucial part of Information Extraction (IE) that involves the detection and classification of named items in unstructured text data. Named entities refer to a broad variety of entities, such as persons, businesses, geographical locations, precise dates, numerical values, and other pertinent categories. Named Entity Recognition (NER) is a prominent area of research that has attracted substantial academic attention and has several practical applications in the field of natural language processing. The use of NER approaches varies across different domains, giving rise to distinct difficulties and possibilities within this subject. Deep learning techniques for NER have demonstrated encouraging outcomes across diverse areas such as healthcare, legal, and chemical fields. There have been proposals to utilise active learning methodologies, specifically Label Transfer Learning (LTP), as a means to mitigate the expenses associated with data annotation in the field of NER. In addition, there are shared projects and benchmarks, such as the ROCLING- 2022 shared task focused on Chinese healthcare NER, which furnish datasets and assessment criteria to facilitate the comparison of diverse NER methodologies. Named Entity Recognition is a specific component within the broader field of Information Extraction that falls within the domain of NLP. The primary objective of this system is to discern and categorise designated items, such as individual names, organisational names, geographical places, dates, and other specific nouns, inside the text that lacks a predetermined framework [2-4]. Named Entity Recognition is an essential preprocessing task in several downstream NLP applications, including but not limited to question answering, conversation systems, and knowledge graph generation [3].
The use of Named Entity Recognition algorithms exhibits variability across many domains, owing to the distinctive characteristics inherent to each specific area [4]. Nevertheless, the procedure often encompasses other essential NLP stages, such as tokenization, part-of-speech tagging, parsing, and model construction [4]. In the field of medicine, for instance, Named Entity Recognition encompasses the task of recognising medical terminology, properties such as negation and severity, and establishing connections between terms and ideas inside domain-specific ontologies [5–9].
In the early stages of development, Named Entity Recognition (NER) systems mostly relied on manually constructed rules, lexicons, and ontologies. However, these systems need extensive dictionaries and meticulous manual feature creation, which constrained their ability to expand and reach maximum performance [10, 11]. In the era of Machine Learning, there are solutions that include machine learning concepts as a potential alternative [12, 13]. However, even these solutions required extensive feature engineering work, limiting their flexibility and effectiveness. The scenario saw significant shifts due to the emergence of the Deep Learning revolution. Recent advancements have brought about advanced deep learning structures like Bi-directional Long Short-Term Memory Networks (BiLSTM) combined with Conditional Random Fields (CRF) and GRAM-CNN [14]. The developments have far beyond the capability of older approaches, demonstrating improved precision and resilience. The greatest significant advancement in Named Entity Recognition (NER) technology occurred with the development of the BERT architecture (Bi-directional Encoder Representations from Transformers). BERT has introduced a new age by using contextual information, allowing NER systems to attain exceptional levels of accuracy and recall [15-18]. The capacity to understand subtle contextual signals has transformed NER accuracy, representing a significant advancement in biomedical text comprehension and data retrieval.
Named Entity Recognition can provide difficulties, particularly in situations involving low-resource languages or topics that necessitate a time-consuming and knowledge-intensive annotation procedure [4, 5, 19]. In order to tackle these issues, scholars have undertaken investigations into many methodologies, including few-shot learning. This particular methodology endeavours to train machine learning models with a very restricted amount of accessible data [19-21]. Furthermore, the integration of entity definition information and domain-specific resources, such as medical dictionaries and ontologies [8, 13, 14, 22], has been shown to enhance the efficacy of Named Entity Recognition models within certain domains [18, 23-32]. Multiple strategies are employed in order to enhance the precision of NER. Several approaches can be identified, including:
1. The process of data augmentation encompasses the generation of supplementary training instances through the modification or synthesis of pre-existing examples. This approach has the potential to enhance the generalisation capabilities of the Named Entity Recognition paradigm [33-36].
2. The hybrid approach involves integrating rule-based methodologies with machine learning techniques, namely Conditional Random Fields (CRF), to enhance the precision of Named Entity Recognition [34, 37, 38].
3. The inclusion of domain-specific resources, such as medical dictionaries and ontologies, together with entity definition information, has been found to enhance the effectiveness of named entity recognition models in certain domains [39-41].
4. Semi-supervised learning is a technique that involves training a model using a little amount of labelled data together with a large amount of unlabeled data. This technique has the potential to improve the effectiveness of named entity recognition models in situations typified by restricted resources, such as low-resource languages or domains where the annotation process requires substantial time, knowledge, and expertise [33, 42, 43]. One way to address the problem of noisy data and the introduction of unwanted patterns during training is to use an iterative training process and data-generating tools. This entails retraining the model using a subset of the original annotated dataset [33, 44, 45].
5. Fine-tuning and transfer learning are often employed techniques in the field of named entity recognition to enhance the efficiency and/or accuracy of NER models [46-48]. Researchers have achieved enhanced precision in NER models across several domains, such as healthcare, legal, and chemical domains, through the use of these methodologies [44, 49]. The job of Named Entity Recognition poses significant challenges in the context of clinical writing, primarily due to the intricate and diverse nature of medical language. Nevertheless, many methodologies may be employed to enhance the precision of Named Entity Recognition in clinical discourse [47].
Several approaches can be identified, including the integration of domain-specific resources, including entity definition information and specialised resources like medical dictionaries and ontologies, which have been shown to enhance the efficacy of named entity recognition models within the medical domain [18, 50]. The hybrid approach involves integrating rule-based methodologies with machine learning techniques, namely CRF, in order to enhance the precision of Named Entity Recognition [51]. Data augmentation is a methodology that entails the creation of supplementary training instances through the alteration or synthesis of preexisting examples. This approach has the potential to enhance the generalisation capabilities of the Named Entity Recognition paradigm [9]. Semi-supervised learning is a methodology that entails training a model using a small quantity of labelled data with a substantial quantity of unlabelled data. This approach has the potential to enhance the efficacy of Named Entity Recognition models in scenarios characterised by limited resources, such as low-resource languages or domains where the annotation process necessitates substantial effort, skill, and domain knowledge [50, 52]. Fine-tuning and transfer learning techniques can be employed to enhance the efficiency and/or accuracy of Named Entity Recognition models [53, 54]. The utilisation of a multilevel named entity recognition framework can effectively tackle the difficulties associated with clinical NER. This framework enables the construction of models that are capable of handling more intricate NER tasks [53, 55]. Through the use of these methodologies, scholars have successfully enhanced the precision of Named Entity Recognition models within the medical field. As an illustration, a research endeavour pertaining to the identification and extraction of adverse drug events from clinical literature by the use of medicine and associated techniques yielded F1 scores of 93.45% for Named Entity Recognition [50]. A recent investigation pertaining to the identification of medication names and their corresponding properties from discharge summaries demonstrated a notable F1-score of 0.91 in the context of the 2018 National NLP Clinical Challenges (n2c2) shared task [50]. Identifying uncommon medical conditions and their associated clinical presentations, together with the influence of socio- economic determinants of health, can provide challenges within the clinical literature. Moreover, the intricate and diverse nature of medical vocabulary presents a formidable obstacle for Named Entity Recognition when applied to clinical material. The concept of stupor poses a challenge in terms of its specific clinical definition [5]. Nevertheless, scholars have put forth various methodologies to enhance the precision of Named Entity Recognition in clinical text [56-59]. These methodologies encompass the integration of domain-specific resources, the adoption of a hybrid approach, data augmentation techniques, semi-supervised learning methods, fine-tuning and transfer learning strategies, as well as the utilisation of a multilevel NER framework [58, 60–62]. Identifying abbreviations and acronyms in clinical material is difficult for Named Entity Recognition algorithms. Nevertheless, many methodologies may be employed to enhance the precision of Named Entity Recognition in clinical discourse containing abbreviations and acronyms. Several approaches can be identified, including the use of domain-specific resources, such as medical dictionaries and ontologies, that enhance the ability of named entity recognition models to identify abbreviations and acronyms in the clinical text [37]. The hybrid approach involves integrating rule-based methodologies with machine learning techniques, namely CRF, in order to enhance the precision of Named Entity Recognition [51]. Data augmentation is a methodology that entails the creation of supplementary training instances through the alteration or synthesis of preexisting examples. This approach has the potential to enhance the generalisation capabilities of the Named Entity Recognition model [55]. Fine-tuning and transfer learning are two techniques that may be employed to enhance the efficiency and/or accuracy of Named Entity Recognition models [53].The utilisation of a multilevel named entity recognition framework presents a viable solution to tackle the obstacles encountered in clinical NER. This framework enables the construction of models that cater to more intricate NER tasks [63].
Through the use of these methodologies, scholars have successfully enhanced the precision of Named Entity Recognition models when applied to clinical material containing abbreviations and acronyms. As an illustration, a research investigation on clinical named entity identification attained an F1-score of 0.91 on the 2018 National NLP Clinical Challenges (n2c2) shared task [27, 50]. When applied to the field of biomedical research, the Biomedical Named Entity Recognition (BioNER) technique is an essential part of the process of biomedical text mining [44, 64-66]. The enormous task of recognising and categorising the numerous things that can be found in biomedical literature is taken on by BioNER. These entities include genes, diseases, chemical names, and medicinal names. The ever-increasing number of biological entities, their plethora of synonyms, the popularity of abbreviations, the use of lengthy entity descriptions, and the combination of letters, symbols, and punctuation within these texts all contribute to this complexity. BioNER, integral to numerous applications in the field of natural language processing, plays a pivotal role in biomedical literature mining [25, 67, 68]. One prominent application involves biomedical relation extraction, which seeks to uncover intricate relationships between diverse biomedical entities, such as diseases, genes, species, and chemicals. The accuracy and quality of downstream relation extraction tasks are directly contingent on the performance of BioNER systems [69]. Furthermore, BioNER significantly contributes to the identification and classification of drugs and their interactions, critical for applications like drug discovery, drug safety monitoring, and personalized medicine [70]. It also facilitates knowledge base completion, enriching biomedical knowledge bases by automatically extracting and categorizing pertinent entities from text. This enriched knowledge is then harnessed for a multitude of purposes, including semantic search, question answering, and data integration. BioNER's utility extends to biomedical question answering, assisting in understanding and responding to complex biomedical queries by identifying and categorizing relevant entities from both the question and the relevant text [71]. In the realm of biomedical text summarization, BioNER proves invaluable in identifying and extracting key biomedical entities from texts, thereby enabling the generation of concise, informative summaries [71]. Moreover, it enhances the accuracy and relevance of search results in biomedical information retrieval systems, where it identifies and classifies pertinent entities in both the query and the document collection [51, 67, 71, 72]. A simplified representation for identifying a Named entity can be represented as shown in Algorithm 1
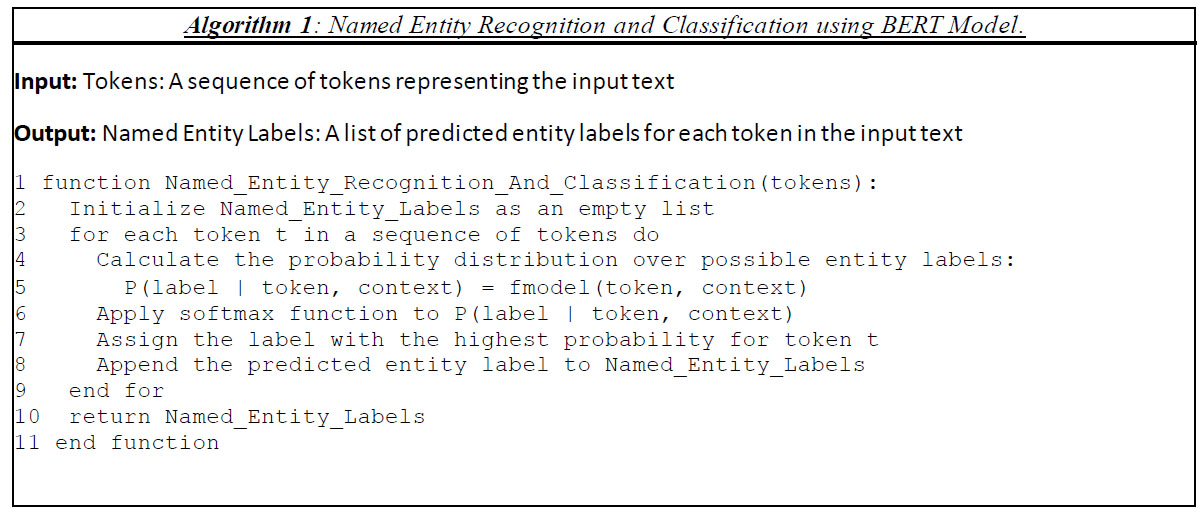
The actual calculation of these probabilities can involve complex machine learning models such as CRFs, BiLSTM-CRFs, or Transformer-based models, which take into account the token, its context, and various features for labelling.
In Algorithm 1:
- P(label | token, context) represents the probability of a specific label given the token and its context.
- fmodel() represents the function that the machine learning model (e.g., CRF, BiLSTM, Transformer) uses to predict this probability.
- The model represents the machine learning model that has been trained to predict entity labels.
SciSpaCy, a Python library and models tailored for practical biomedical and scientific text processing, makes extensive use of spaCy [73]. spaCy itself employs deep learning techniques and, with the release of version 3, has transitioned to using the Transformer architecture as its deep learning model. It offers a customizable architecture that accommodates various deep learning techniques such as LSTMs, CRFs, and transformers to suit specific tasks [74].
BioBERT, on the other hand, is a pre-trained biomedical language representation model based on the BERT architecture [75]. It has been fine-tuned on an extensive corpus of biomedical literature, incorporating articles from PubMed and PMC as shown in Fig. (1). The adaptability of this model enables it to perform very well in many biomedical activities, such as biomedical named entity identification, connection extraction, question answering, and drug discovery. Both BioBERT and SciSpaCy are open-source tools, rendering them important resources for NER.
In this work, we present significant contributions to the domain of Biomedical Named Entity Recognition (BioNER), with prior focus on the utilization and examination of two prominent models, SciSpaCy and BioBERT, addressing the intricate challenges inherent in BioNER.
1. Through meticulous training on diverse biological datasets, we systematically evaluate the performance of SciSpaCy and BioBERT across various domains. Our comprehensive assessment employs key metrics such as F1 scores and processing speed, providing a detailed understanding of their efficacy in BioNER tasks.
2. The study offers valuable insights for the selection of tools tailored to specific BioNER requirements. By optimizing Named Entity Recognition in the field of biomedical research, our work aims to contribute to the broader advancement of this critical domain
We have performed some literature surveys of the papers that collectively discuss various approaches to biomedical NER. Amith 2017 proposes an ontology-driven method that utilizes information extraction and features of ontologies to identify biomedical software names [76]. Al-Hegami 2017 focuses on the use of machine learning classifiers and a rich feature set to recognize biomedical entities, with the K-Nearest Neighbour classifier showing promising results [77]. Zhang 2013 presents an unsupervised approach to NER, using a noun phrase chunker and distributional semantics for entity extraction and classification [20].
Li 2023 proposes a fusion multi-features embedding method, combining deep contextual word-level features, local char-level features, and part-of-speech features to improve biomedical entity recognition [78]. Kaewphan et al., 2018, published in the “Database Journal of Biological Databases Curation,” shows a system for automatically identifying a wide range of biomedical entities within literature, boasting state-of-the-art performance. The study achieved the highest results in named entity recognition, underscoring its capability in biomedical entity recognition and normalization [79].
Priyanka et al., 2021 provided a detailed survey of clinical Named Entity Recognition and Relationship Extraction (RE) techniques, addressing existing NLP models, performance, challenges, and future directions in the context of information extraction from clinical text, offering insights on current research and evaluation metrics [18].
Gong et al., 2009 published in the “International Conference on BioMedical Engineering and Informatics,” this study introduces a hybrid approach aimed at recognizing untagged biomedical entities. The approach demonstrates its utility in identifying biomedical entities in the GENIA 3.02 corpus, offering support for biologists in tagging biomedical entities within biomedical literature [80].
Kanimozhi & Manjula in 2018 performed a systematic review that focuses on the identification and classification of named entities within unstructured text documents. The goal is to extract valuable information from such texts by identifying and categorizing the named entities, a crucial step in extracting useful knowledge from unstructured textual data [81].
Overall, these papers highlight different techniques and algorithms for biomedical NER, showcasing the importance of effective natural language processing tools in organizing and extracting information from biomedical literature. Table 1 depicts the core methodology of the above-mentioned research works.
| S. No | Title | Authors | Year | Methodology |
|---|---|---|---|---|
| 1. | “Knowledge-Based Approach for Named Entity Recognition in Biomedical Literature: A Use Case in Biomedical Software Identification” | Muhammad Amith, et al. [76] | 2017 | Ontology-driven method to identify familiar and unfamiliar software names. |
| 2. | “A Biomedical Named Entity Recognition Using Machine Learning Classifiers and Rich Feature Set” | A. S. Al-Hegami, et al. [77] | 2017 | K-nearest neighbor trained with suitable features for recognizing biomedical named entities. |
| 3. | “Unsupervised biomedical named entity recognition: Experiments with clinical and biological texts” | Shaodian Zhang, et al. [20] | 2013 | Noun phrase chunker followed by a filter based on inverse document frequency for candidate entity extraction. |
| 4. | “Biomedical named entity recognition based on fusion multi-features embedding” | Meijing Li, et al. [78] | 2023 | A proposed multi-feature embedding method with a positive effect on prediction results. |
| 5. | “Wide-scope biomedical named entity recognition and normalization with CRFs, fuzzy matching and character level modelling” | S. Kaewphan, et al. [79] | 2018 | System for automatically identifying a multitude of biomedical entities from the literature with state-of-the-art performance. |
| 6. | “A Survey on Recent Named Entity Recognition and Relationship Extraction Techniques on Clinical Texts” | Priyanka et al. [18], | 2021 | Surveyed clinical Named Entity Recognition and Relationship Extraction (RE) techniques and addressed existing NLP models, performance, challenges. |
| 7. | “A Hybrid Approach for Biomedical Entity Name Recognition” | Lejun Gong, et al. [80] | 2009 | A hybrid approach for recognizing untagged biomedical entities in GENIA 3.02 corpus. |
| 8. | “A Systematic Review on Biomedical Named Entity Recognition” | U. Kanimozhi, et al. [81] | 2018 | Focus on identifying and classifying named entities for extracting useful information from unstructured text documents. |
In addition to this study, there are several domains where Named Entity Recognition may be applied. Potential intersections and possibilities for collaboration or cross-pollination exist across the fields, especially in areas such as Visual Named Entity Recognition, cross-modal learning, semantic comprehension, and multi-modal information fusion.
1. Visual Named Entity Recognition (VNER): is a process that involves detecting and classifying named entities in text-based data. There is an increasing interest in Visual Named Entity Recognition, which refers to the process of recognising and categorising named items in photos or videos. The domain might possibly benefit from the application or adaptation of image processing techniques, particularly those that emphasise feature extraction and semantic interpretation.
2. Cross-Modal Learning: Cross-modal learning involves leveraging information from different modalities (e.g., text and images) to improve performance on various tasks. Techniques developed for image processing, especially those involving attention mechanisms and feature extraction, could potentially be integrated with NER systems to improve entity recognition performance, especially in cases where textual context alone may not be sufficient.
3. Semantic Understanding: Both image processing and NER involve understanding the semantic content of data, albeit in different modalities. Techniques developed for image inpainting, super-resolution, or feature extraction may enhance the semantic understanding of textual data, which could indirectly benefit NER systems by providing richer contextual information.
4. Multi-Modal Information Fusion: Another potential linkage is through multi-modal information fusion techniques. Research in image processing often involves integrating information from different sources or modalities to improve the quality of image reconstruction or understanding. Similarly, integrating information from images (e.g., scenes, objects) with textual data (e.g., context, descriptions) could enhance NER systems' performance, especially in scenarios where textual and visual information are both available.
Some interesting works where NER can be combined are done by Yuantao Chen et al. in image processing techniques such as image inpainting and super-resolution. In a series of innovative contributions by Yuantao Chen, Runlong Xia, Kai Yang, and Ke Zou, their research endeavors have significantly advanced the field of image processing and restoration. In June 2023, they introduced “DARGS Image inpainting algorithm via deep attention residuals group and semantics,” proposing a method that combines Semantic Priors Network, Deep Attention Residual Group, and Full-scale Skip Connection to effectively restore missing regions in images. Building on this foundation [82], in October 2023, they presented “GCAM: lightweight image inpainting via group convolution and attention mechanism,” offering a lightweight approach that utilizes group convolution and rotating attention mechanism to enhance image inpainting while optimizing resource usage [83]. In January 2024, they unveiled “MFMAM: Image inpainting via multi-scale feature module with attention module,” introducing a network utilizing multi-scale feature module and improved attention mechanisms for enhanced image inpainting, particularly focusing on texture and semantic detail preservation [84]. Their work continued with “DNNAM: Image inpainting algorithm via deep neural networks and attention mechanism,” published in March 2024, where they introduced a method incorporating partial multi-scale channel attention mechanism and deep neural networks to improve image inpainting accuracy and quality [85]. Lastly, in July 2024, their paper titled “MICU: Image super-resolution via multi-level information compensation and U-net” presented a novel approach for image super-resolution using multi-level information compensation and U-net architecture, significantly improving image reconstruction quality compared to existing methods [86]. Collectively, these papers underscore the author group's commitment to advancing image processing techniques with innovative methodologies and improved performance metrics. These works are very close in the possibilities for collaborating NER in areas such as Visual Named Entity Recognition, cross-modal learning, semantic comprehension, and multi-modal information fusion.
2. METHODOLOGY
The work aimed to rigorously evaluate the performance of BioBERT and SCISpaCy in biomedical Named Entity Recognition, considering the data's origin, preprocessing, training, and evaluation processes. The choice of these models and the training process was guided by the aim of achieving accurate and generalizable recognition of biomedical entities. These considerations are explained as follows:
2.1. Data Collection
The dataset utilized in this study was curated from diverse sources of biomedical Named Entity Recognition datasets. These datasets, including JNLPBA [15], BC4_CHEMD [87], BIONLP13CG [88], EXPERT 206 [70], and BC5CDR [89], were selected to encompass a wide range of biomedical entity types and domains, offering a comprehensive evaluation of BioNER models as shown in Table 2.
Entity Types and Datasets: The evaluation encompassed multiple datasets, each representing distinct biomedical entity types. The datasets included.
- JNLPBA: Targeted entities included DNA, CELL_TYPE, CELL_LINE, RNA, and PROTEIN.
- BC4_CHEMD: Focused on the CHEMICAL entity type.
- BIONLP13CG: Encompassed a diverse set of entity types, such as AMINO_ACID, ANATOMICAL_SYSTEM, CANCER, and more.
- EXPERT 206: Included GENE, DISEASE, and VARIANT as recognized entities.
- BC5CDR: Considered both CHEMICAL and DISEASE entities.
2.2. Data Preprocessing
2.2.1. Data Conversion for BioBERT
To prepare the data for training BioBERT, the datasets were converted into a BIO (Beginning-Inside-Outside) format. This involved annotating each token in the dataset with one of three labels: “B-” for the beginning of an entity, “I-” for internal parts of an entity, and “O” for tokens outside of any entity. This format enables BioBERT to understand and recognize the boundaries of biomedical entities within the text.
2.2.2. Data Conversion for SCISpaCy
For training SCISpaCy, the datasets were transformed into a format suitable for spaCy's NER model. The conversion process required structuring the data as a list of sentences, each followed by a list of (start, end, label) triples, where the triples define the start and end positions of the entity within the sentence and its associated label. For example, (“Tokyo Tower is 333m tall.”, [(0, 11, “BUILDING”)]) represents a sentence, the start and end positions of the entity “BUILDING,” and its label.
2.3. Data Splitting
To facilitate model evaluation, the dataset was divided into training and testing subsets. The data was partitioned using an 80-20 split, allocating 80% of the data for training and 20% for testing. This ensures that the models are trained on a substantial portion of the data while preserving an independent dataset for assessing their generalization performance.
2.4. Hardware and Software Environment
Training of the model was carried out on carefully managed hardware consisting of Google Collab T4 GPUs. The environment had the essential programming languages, libraries, and frameworks for the construction and execution of the models, as well as particular GPU specifications for rapid training.
| Datasets | Entity Type | No. of Entities |
|---|---|---|
| BIONLP13CG | Tissues | 587 |
| BIONLP13CG | Organisms | 2,093 |
| BIONLP13CG | Gene/Protein | 7,908 |
| EXPERT 206 | Disease | 7,390 |
| EXPERT 206 | Gene/Protein | 7,393 |
| BC5CDR | Disease | 12,204 |
| BC5CDR | Chemical | 12,204 |
| BC4_CHEMD | Chemical | 84,249 |
| JNLPBA | Cell Line | 4,315 |
Table 3.
| Model | Entity Types | SCISpaCy F1 Score | BioBERT F1 Score |
|---|---|---|---|
| JNLPBA | “DNA, CELL_TYPE, CELL_LINE, RNA, PROTEIN” | 73.1 | 73.67 |
| BC4_CHEMD | “CHEMICAL” | 85.1 | 86.07 |
| BIONLP13CG | “AMINO_ACID, ANATOMICAL_SYSTEM, CANCER, CELL, CELLULAR_COMPONENT, DEVELOPING_ANATOMICAL_STRUCTURE, GENE_OR_GENE_PRODUCT, IMMATERIAL_ANATOMICAL_ENTITY, MULTI-TISSUE_STRUCTURE, ORGAN, ORGANISM, ORGANISM_SUBDIVISION, ORGANISM_SUBSTANCE, PATHOLOGICAL_FORMATION, SIMPLE_CHEMICAL, TISSUE” | 78.13 | 86.07 |
| EXPERT 206 | “GENE, DISEASE, VARIANT” | 91.08 | 91.23 |
| BC5CDR | “CHEMICAL, DISEASE” | 85.53 | 87.83 |
3. RESULTS AND DISCUSSION
3.1. Model Performance
3.1.1. JNLPBA Dataset
For the JNLPBA dataset, SCISpaCy and BioBERT both displayed competitive performance, with F1 scores of 73.1 and 73.67, respectively. These ratings were uniform across all entity types in this sample.
In the BC4 CHEMD dataset, which concentrates on CHEMICAL items, both models demonstrated excellent performance. BioBERT surpassed SCISpaCy with an F1 score of 86.07, while SCISpaCy received an F1 score of 85.1.
3.1.2. BIONLP13CG Dataset
The BIONLP13CG dataset featured a large variety of entity types, posing a challenge to the models' ability to recognise a wide range of biomedical items. BioBERT's F1 score of 86.07 was superior to SCISpaCy's score of 78.13.
The EXPERT 206 dataset, which consists of GENE, DISEASE, and VARIANT entities, produced high F1 scores for both models. SCISpaCy obtained an F1 value of 91.08, but BioBERT fared somewhat better with an F1 score of 91.23.
3.1.3. BC5CDR Dataset
For the BC5CDR dataset, which involves CHEMICAL and DISEASE entity recognition, both models displayed strong performance. SCISpaCy achieved an F1 score of 85.53, while BioBERT exhibited superior performance with an F1 score of 87.83. These results are showcased in Table 3.
3.2. Inference and Performance Trade-offs
The evaluation of inference times revealed that SCISpaCy processed an average sentence in approximately 90 milliseconds, while BioBERT required an average of 278 milliseconds per sentence as shown in Fig. (2). It is important to note that the relatively longer inference time of BioBERT can be attributed to various internal tasks, including tokenizer loading and token classification.
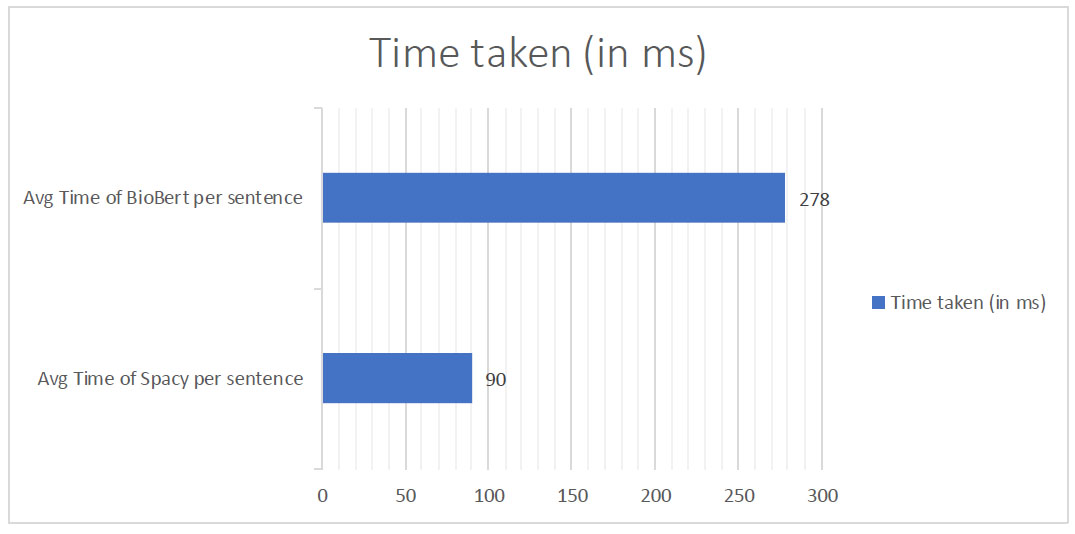
However, BioBERT compensates for this by efficiently processing multiple sentences in parallel. This capability enhances throughput, making BioBERT a favorable choice in scenarios where parallelism is crucial.
4. DISCUSSION
Based on the results showcased in Table 3. Some inferences have been deduced. These inferences can help to guide the selection of the appropriate model for specific biomedical NER tasks based on the dataset's entity types and performance requirements.
4.1. Entity Types and Specialization
The F1 scores vary across different entity types and datasets. For instance, in the JNLPBA dataset, DNA, CELL_TYPE, CELL_LINE, RNA, and PROTEIN have relatively similar F1 scores, indicating that the model's performance is consistent across these biomedical entity types. On the other hand, in the BIONLP13CG dataset, there is a wide range of entity types with varying F1 scores, suggesting that the model's performance is more specialized for some entity types.
4.2. BioBERT's Consistency
BioBERT consistently achieves higher F1 scores compared to SCISpaCy across all datasets and entity types. This indicates BioBERT's robustness and effectiveness in biomedical named entity recognition tasks.
4.3. High Performance for EXPERT 206
The EXPERT 206 dataset demonstrates the highest F1 scores for both models. This suggests that the entities in this dataset, such as Genes, Diseases, and variants, are well-recognized by both SCISpaCy and BioBERT.
4.4. Strong BioBERT Performance in BC5CDR
In the BC5CDR dataset, which focuses on Chemicals, BioBERT significantly outperforms SCISpaCy with an F1 score of 87.83, indicating its strength in recognizing chemical entities.
4.5. Balanced Performance in BC4_CHEMD
The BC4_CHEMD dataset, consisting of DISEASE and CHEMICAL entity types, shows similar and high F1 scores for both models. This suggests that both SCISpaCy and BioBERT are proficient in recognizing these types of entities.
4.6. Biomedical Specialization
The choice of model may depend on the specific biomedical entity types in the dataset. While BioBERT generally outperforms SCISpaCy, practitioners should consider the nature of the entities they are dealing with when selecting the most suitable model for their task.
4.7. Overall Robustness
Both SCISpaCy and BioBERT demonstrate strong performance in biomedical named entity recognition. However, BioBERT's consistently higher F1 scores and adaptability to various entity types make it a valuable choice for a wide range of biomedical applications.
4.8. Time Complexity on Same Hardware
On the same hardware configuration with a T4 GPU (utilizing Google Colab's default GPU settings), we observed that spaCy demonstrated a faster average inference time per sentence, approximately 50 milliseconds, compared to BioBERT's average speed of 178 milliseconds per sentence. It is worth noting that the apparent time difference in favor of spaCy may be attributed to its streamlined processing for individual sentences. However, it is important to consider that BioBERT exhibits a significant advantage when processing multiple sentences in parallel. This capability compensates for the additional time incurred by BioBERT in various internal tasks such as tokenizer loading and token classification. Consequently, BioBERT's suitability for parallel sentence processing makes it a strong contender in scenarios where high throughput and parallelism are critical.
CONCLUSION
SCISpaCy and BioBERT, along with other BioNER models, have demonstrated their capabilities, benefits, and relevant considerations. This study evaluated diverse datasets, and performance metrics, offering valuable insights to scholars and practitioners in the field of biological natural language processing. The study produced intriguing findings. The selection of a model such as SCISpaCy or BioBERT significantly impacts the detection of many biological events. BioBERT consistently achieved higher F1 scores than SCISpaCy across several datasets, entity types, and domains. The durability and adaptability of BioBERT in the biomedical field make it a highly promising choice for BioNER.The datasets exhibited notable variations in F1 scores across different item categories. The F1 ratings for both models in the EXPERT 206 dataset of Genes, Diseases, and Variants indicate that SCISpaCy and BioBERT effectively recognise and categorise these entity types. The BIONLP13CG dataset exhibited a diverse range of entity kinds, each with distinct F1 scores. The performance of the model may be tailored to specific entities. Parallel processing is an additional advantage of BioBERT. Although BioBERT may have longer phrase inference times, it has the ability to process several sentences simultaneously, which makes it an excellent choice for high-throughput applications. It is essential to assess the different categories within the dataset and determine the optimal trade-off between efficiency and precision when recognising biological entities. The selection of the model should be guided by the intrinsic aspects of the work. Our analysis demonstrates the significance of BioNER in the field of biomedical research and the capabilities of current models. Before making a decision, it is important to thoroughly comprehend the specific criteria and objectives of each biological Named Entity Recognition task, and then select either SCISpaCy, BioBERT, or a combination of both accordingly. The advancement of NLP models in the field of biomedicine provides professionals with a diverse array of tools to effectively extract pertinent information from texts related to biology.
LIMITATIONS, FUTURE WORK AND RESEARCH DIRECTIONS
To improve the thoroughness of our assessments, it is crucial to further investigate domain-specific algorithms for Named Entity Recognition in the context of future studies. In order to achieve this objective, we suggest a deliberate enlargement of our study scope to encompass notable algorithms such as Bert-PKD, CollaboNet, TinyBert, BERN2, and other state-of-the-art techniques. Integrating these algorithms into our future endeavours will enhance the strength and comprehensiveness of our evaluation system. The limitations in the field of Biomedical Named entity recognition include data availability, expert annotation requirements, and the vast space of biomedical concepts. Our objective is to analyse a wider range of NER algorithms in a systematic manner, in order to reveal subtle variations in performance and find methods that may have distinct benefits in particular biological scenarios. This strategic growth is in line with our dedication to developing the area of Biomedical Named Entity Recognition and guaranteeing that our research offers practical insights for researchers, practitioners, and professionals in the biomedical sector. Integrating new algorithms will enhance the comprehensiveness of our studies and promote a more nuanced comprehension of the advantages and constraints of different techniques. To summarise, more study on domain-specific algorithms for NER is crucial. There are potential intersections and opportunities for collaboration or cross-pollination between the fields, particularly in areas such as Visual Named Entity Recognition, cross-modal learning, semantic understanding, and multi-modal information fusion. We are committed to expanding the reach and significance of our findings in future studies.


