RESEARCH ARTICLE
Classification of Colorectal Cancer using ResNet and EfficientNet Models
Abhishek1, Abhishek Ranjan1, Priyanshu Srivastva1, B Prabadevi1, *, Sivakumar Rajagopal2, Rahul Soangra3, Shamala K. Subramaniam4
Article Information
Identifiers and Pagination:
Year: 2024Volume: 18
E-location ID: e18741207280703
Publisher ID: e18741207280703
DOI: 10.2174/0118741207280703240111075752
Article History:
Received Date: 21/09/2023Revision Received Date: 27/11/2023
Acceptance Date: 27/12/2023
Electronic publication date: 16/01/2024
Collection year: 2024

open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution 4.0 International Public License (CC-BY 4.0), a copy of which is available at: https://creativecommons.org/licenses/by/4.0/legalcode. This license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Introduction
Cancer is one of the most prevalent diseases from children to elderly adults. This will be deadly if not detected at an earlier stage of the cancerous cell formation, thereby increasing the mortality rate. One such cancer is colorectal cancer, caused due to abnormal growth in the rectum or colon. Early screening of colorectal cancer helps to identify these abnormal growth and can exterminate them before they turn into cancerous cells.
Aim
Therefore, this study aims to develop a robust and efficient classification system for colorectal cancer through Convolutional Neural Networks (CNNs) on histological images.
Methods
Despite challenges in optimizing model architectures, the improved CNN models like ResNet34 and EfficientNet34 could enhance Colorectal Cancer classification accuracy and efficiency, aiding doctors in early detection and diagnosis, ultimately leading to better patient outcomes.
Results
ResNet34 outperforms the EfficientNet34.
Conclusion
The results are compared with other models in the literature, and ResNet34 outperforms all the other models.
1. INTRODUCTION
Colorectal cancer, also known as bowel or colon cancer, is a malignant illness originating in the colon or rectum. It is caused by uncontrolled proliferation of aberrant cells in the large intestine lining. Symptoms include changes in bowel habits, stool blood, stomach pain, weight loss, and exhaustion. Early diagnosis and prompt medical intervention are crucial for effective treatment.
Colorectal Cancer is a major global cancer causetherefore, early detection is crucial for prompt management and improved patient outcomes. Deep learning techniques, particularly CNNs, have shown promise in medical imaging applications, including histological image categorization for cancer diagnosis. Optimizing CNNs for colorectal cancer classification using histological images is a critical research issue [1]. Recently, deep learning has played a vital role in almost all applications and has brought remarkable results in those applications [2].
Improved deep-learning CNNs are needed for colorectal cancer categorization. Early identification is crucial for effective control and treatment. Some attempts to detect various cancers at an early stage were carried out using machine learning algorithms [3]. Despite advancements in medical imaging and histology, detecting colorectal cancer using histological images remains challenging due to subjectivity, human interpretations, and inter-observer heterogeneity among pathologists. This can affect treatment recommendations and require manual effort, which can be time-consuming and not scalable for addressing the growing number of cases [4]. Histological scans offer microscopic insights, but human interpretation takes time and is prone to diagnostic mistakes. Therefore, automated systems are needed to accurately classify colorectal cancer from histological images, aiding pathologists in their diagnosis process.
CAD systems for Colorectal Cancer detection often face limitations in feature extraction and representation, leading to inferior classification accuracy. Handcrafted features may not accurately represent subtle patterns in histology pictures, and models lack depth and complexity for extracting high-level characteristics, limiting their discriminating capabilities.
Optimizing deep-learning CNNs for colorectal cancer classification using histological images is crucial for improving patient outcomes, automating categorization processes, and revolutionizing cancer diagnosis. By boosting accuracy and efficiency, early intervention and personalized treatment strategies can be implemented, while automation reduces pathologist workload and potentially lowers diagnostic disparities. Advanced deep learning algorithms can also revolutionize cancer diagnosis, enabling more complex AI-assisted medical applications.
The paper is organized as follows: Section 2 provides details of various studies in the literature. Section 3 explores the data used and Section 4 details the flow of the study, Section 5 discusses the experimental results, and Section 6 concludes the paper.
2. MATERIALS AND METHOD
2.1. Literature Surveys
This section reviews various studies on deep learning and machine learning techniques for colorectal cancer detection and classification. Topics covered include image-based classification, predictive models, vision transformers, hybrid deep learning frameworks, and feature engineering and transfer learning approaches. These studies aim to improve colorectal cancer diagnosis and prognosis accuracy and efficiency while acknowledging challenges and possibilities in the field.
In studies conducted in 2021, Sarwinda et al. utilized variants of networks (ResNet) to detect colorectal cancer [1]. There has been a focus on utilizing deep learning algorithms to categorize colorectal cancer. For that, Ponzio and colleagues introduced a method in 2018 that involved employing networks [4]. Additionally, Tamang and Kim reviewed approaches involving learning methods [5]. Following their work, Bychkov et al. utilized learning-based tissue analysis to predict outcomes related to cancer [6]. Recently, Zeid et al. applied vision transformers for classifying multiclass histology images related to colorectal cancer [7] and Xu et al. proposed a system in 2020 that incorporated learning for the diagnosis of colorectal cancer [8].
In their research, Damkliang et al. utilized deep learning and machine learning techniques to classify types of tissues [9]. Tsai and Tao also employed learning techniques for the classification of colorectal cancer tissues [10]. Ben and Hamida et al. focused, on using learning for the analysis of images related to colon cancer [11]. In 2022, Talukder and colleagues utilized machine learning to detect lung and colon cancer [12]. Pataki et al. made improvements in colorectal cancer screening [13], while Kather et al. explored the use of learning to predict survival rates based on histology cancer slides in 2019 [14].
Moving on to 2023, Khazaee Fadafen and Rezaee proposed a learning framework for classifying colorectal cancer histology images across multiple tissues [15]. Alboaneen et al. discussed the challenges and opportunities associated with predicting cancer using machine and deep learning algorithms [16], while Tsai and Tao provided valuable insights as well [17]. Zhou et al. developed a system called HCCANet [18]. Meanwhile, Hu et al. introduced a dataset called Enteroscope Biopsy Histopathological H&E Image Dataset specifically designed for evaluating image classification in this field [19]. Irawati et al. compared architectures of neural networks [20]
Tasnim et al. developed a model using learning, specifically convolutional neural networks (CNN), for colon cancer patients’ prognosis [21]. Trivizakis et al. proposed a framework called pathomics that aims to classify cancer cases [22], and Kumar et al. Studied techniques for extracting handcrafted and dense features [23]. Lastly, Ohata et al. introduced an approach based on transfer learning for categorizing cancer cases [24].
Masud et al., on the other hand, utilized a strategy based on machine learning to diagnose lung and colon cancers [25]. Also, a framework for detecting lung and colon cancers was employed using machine learning, which classifies different types of lung and colon cancerous cells [27]. Additionally, researchers have made advancements in the recognition of cancer using machine learning techniques using pre-trained models like YOLO and modified R-CNN employed for polyp detection [28, 29].
The survey of the above-mentioned literature on deep learning and machine learning techniques in colorectal cancer detection and classification reveals advances and challenges. Deep neural networks, CNN, ResNet, and attention processes are used to produce accurate models. However, challenges like interpretability, dataset variety, and external validation remain. Overall, promising results are achieved, but limitations remain.
Studies emphasize the potential of deep learning in colorectal cancer detection and prognosis but still need further investigation on comparisons and repeatability across multiple datasets. Transfer learning and feature engineering are feasible tactics. However, overcoming hurdles and overcoming challenges are crucial for practical applicability.
The study presents a unique technique for colorectal cancer classification, utilizing the RESNet34 and EfficientNetB4 architectures. It aims to improve the detection of colorectal cancer by focusing on their complementing characteristics. The research uses the ADAM optimizer for RESNet34 and ADAMmax for EfficientNetB4, demonstrating a sophisticated understanding of the optimization environment for each model. The study also employs a unique set of parameters for model training, including a learning rate of 0.001, batch size of 32, and training for 50 epochs, to balance model convergence and overfitting avoidance. This approach sets the study apart from previous work and provides a new perspective on the ideal training regime for colorectal cancer classification. The research contrasts its selected approaches with those used in foundational publications on colorectal cancer detection, highlighting the unique architectural decisions, optimization methodologies, and hyperparameter setups of the offered models. The experimental data and analysis provide a detailed examination of the proposed models, offering insights into areas for improvement and potential future approaches for colorectal cancer categorization research.
3. SYSTEM SPECIFICATION
3.1. Computing Environment
We utilized the Kaggle Notebook to implement our model for our study. Kaggle offers a cloud-based platform with a variety of resources to meet a variety of computational demands.
- CPU: We utilize two cores, although a quad-core or greater is suggested for intensive computations.
- RAM: 8GB minimum, 16GB or more recommended for memory-intensive activities.
- GPU: A dedicated GPU (e.g., NVIDIA GeForce or Tesla series) can dramatically speed deep learning and other GPU-accelerated processes if suitable.
3.2. Software Specification
Use the Kaggle operating system, which is a Linux-based environment.
- Python Libraries: - Make use of necessary libraries for data processing (Pandas), numerical computation (NumPy), and data visualization (Matplotlib, Seaborn).
- Scikit-Learn and TensorFlow are two machine learning libraries.
- Jupyter Notebooks: Based on Jupyter, Kaggle Notebooks provide an interactive and sharing environment for code execution and documentation.
- Version Control: Use Git for version control to monitor changes and collaborate effectively.
- Data Access: Using data privacy and use rights, access datasets directly from Kaggle's dataset repository.
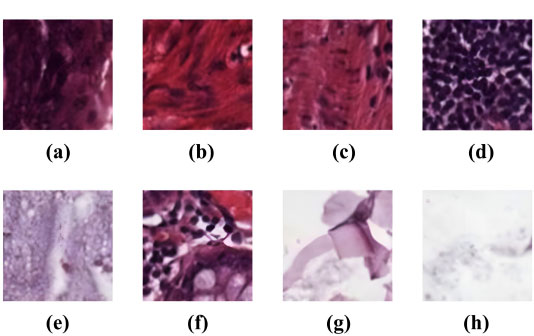 |
Fig. (1a-h). 8 different classes of tissues inside dataset. |
4. EXPLORATORY DATA ANALYSIS
The study utilized the “Kather-texture-2016-image” histology dataset [30]. It consists of 5000 histological images, as depicted in Fig. (1), of human colorectal cancer tissue and healthy normal tissue images. Inside the dataset, each image is 150×150 pixels distributed across eight distinct tissue classes based on their texture characteristics. Over this dataset, the improved CNN model’s architecture was validated for accuracy.
The eight distinct tissue classes are described as follows:
5. EXPERIMENTAL STUDY
Deep learning gained prominence in scientific computing because of CNN and is now frequently employed by businesses to solve complicated problems. We used two CNN models for our research: ResNet34 and EfficientNetB4.
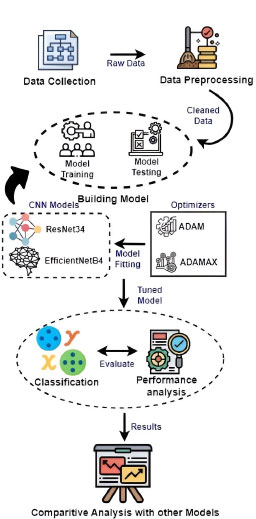 |
Fig. (2). Flow of the proposed research. |
ResNet34 and EfficientNetB4 improve image classification methods for colorectal cancer by mitigating the vanishing gradient problem and training deeper networks. ResNet34's deep residual connections enable effective training of deeper networks, while Efficient NetB4 uses compound scaling for superior representation learning and improved performance on restricted datasets. The architecture's improved design enables greater feature extraction from complex tissue structures, making ResNet34 and EfficientNetB4 viable options for accurate and discriminative cancer classification.
The recognition process in Fig. (2) involves two stages: (1) Model Training and (2) Model Testing.
5.1. Model Training
In experiment 1, the dataset was methodically partitioned, with 80% designated for training and 20% designated for testing. ResNet34, a Convolutional Neural Network (CNN) noted for its success in deep learning tasks, was chosen as the neural network architecture for training. The learning rate was set at 0.005, which is a crucial hyperparameter controlling the step size during optimization and hence influencing the model's convergence and stability.
The Adaptive Moment Estimation (Adam) optimizer was utilized throughout the training phase, which is a widely known approach that changes learning rates for each parameter separately by integrating momentum and RMSprop techniques. The network topologies' performance was carefully investigated, offering a comprehensive view of the model's behavior during the duration of training epochs.
The training configuration included 50 epochs, allowing the model to learn from the dataset repeatedly. Early stopping with a stop_patience of three epochs was adopted to improve training efficiency and reduce overfitting. This method guaranteed that if there was no change in performance on a validation set, the training process was terminated, eliminating excessive iterations that may lead to overfitting.
Mini-batch training was used, using 32 photos in each epoch. When compared to batch gradient descent, this method allows for more computationally efficient model updates. This experiment's results, shown in Fig. (3), give a thorough overview of the model's accuracy or other performance measures over the training epochs, revealing information on its learning dynamics and potential areas for further modification.
In experiment 2, the dataset was methodically separated into three subsets, with 70% going to training, 15% going to validation, and the remaining 15% going to testing. This partitioning technique allows the model to learn from a significant percentage of the data during training, while separate validation and test sets allow for hyperparameter fine-tuning and a full assessment of the final model's performance, respectively.
The experiment's findings were heavily influenced by hyperparameter adjustment. The learning rate, a critical parameter impacting model convergence, was set to 0.001. This lower learning rate is frequently selected for its capacity to contribute to sustained convergence, which is especially important when working with deep neural networks. To guide the optimization process, the Adaptive Moment Estimation with Maximum (Adamax) optimizer was used. Adamax, a variation of the well-known Adam optimizer, is ideal for models with enormous parameter spaces.
 |
Fig. (3). ResNet34 loss vs. learning rate at 0.005. |
The model was evaluated across 50 training epochs, enabling it to loop over the whole training dataset numerous times. As a regularization strategy, early halting with a patience level of three epochs was used. This method prevents overfitting by stopping the training process after three consecutive epochs of no increase in the chosen performance metric on the validation set. Each training epoch's mini-batch size was set to 32 photos, as is customary in deep learning. This mini-batch size strikes a compromise between computing efficiency and taking advantage of parallel processing capabilities.
Figs. (4 and 5) show the results of the experiment, which give visual insights into the model's performance indicators during the training process. These visualizations provide a thorough insight into how the model grows over time, offering light on its learning dynamics as well as the efficiency of the chosen hyperparameters and network design.
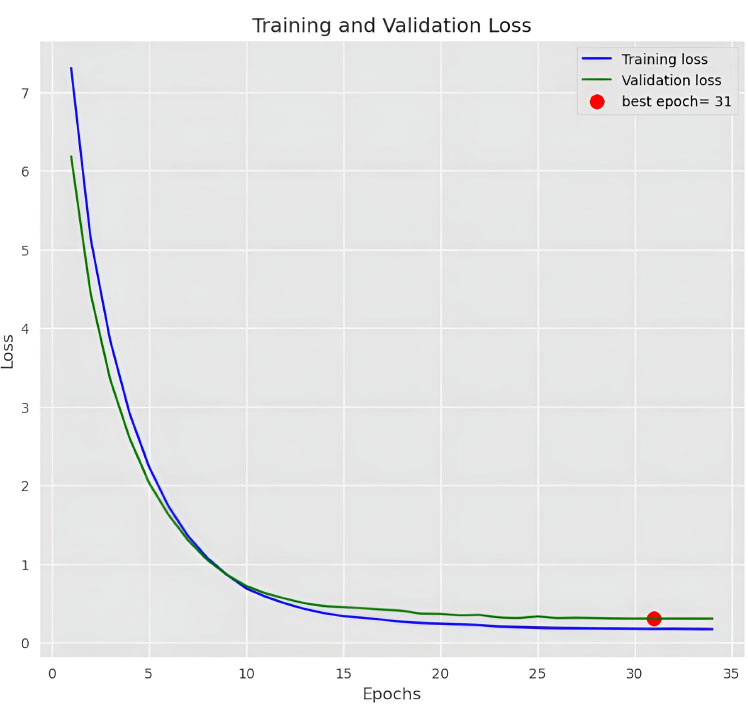 |
Fig. (4). EfficientNetB4 training and validation loss. |
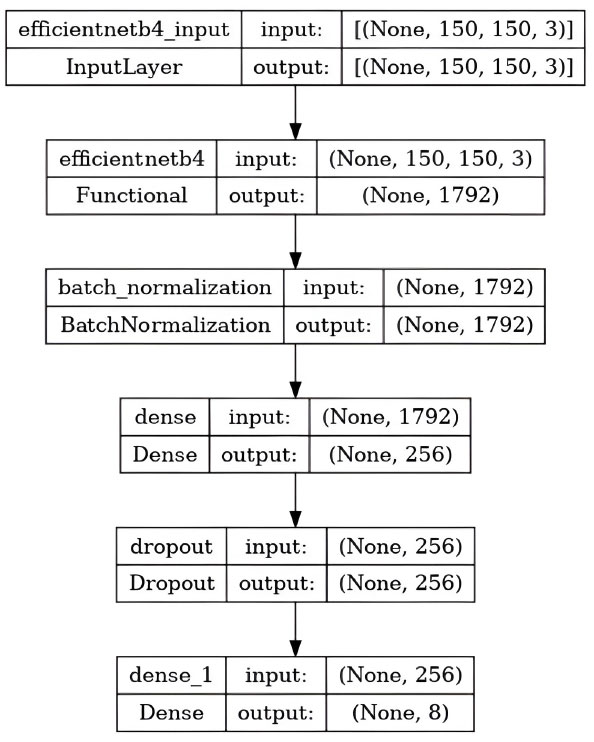 |
Fig. (5). EfficientNetB4 processing illustration. |
5.2. Model Testing
During the model testing phase of our experimental research on the classification of colorectal cancer using CNN models, namely RESNet34 and EfficientNetB4, we did a complete review. The dataset, which was initially divided into training and testing sets, aided in the independent evaluation of the models' generalization ability. We began the testing procedure by loading pre-trained RESNet34 and EfficientNetB4 models and focusing on accuracy as the major assessment criterion.
To guarantee compatibility with the models, histology images from the testing set were meticulously pre-processed. This includes shrinking photos to match the model input size and normalizing pixel values. The models were then analysed using a confusion matrix, which is a useful technique that provides a comprehensive analysis of true positives, true negatives, false positives, and false negatives for each class.
 |
Fig. (6). ResNet34 confusion matrix. |
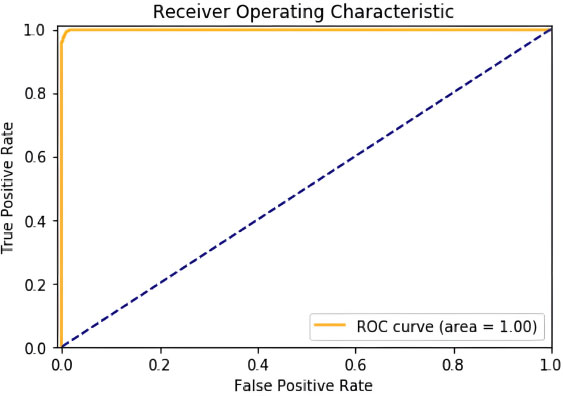 |
Fig. (7). ResNet34 ROC (accuracy). |
The key performance criterion studied was precision, which is critical in the medical setting of colorectal cancer categorization. Precision was calculated for each class by dividing the number of true positive predictions by the total number of true positives and false positives. This resulted in precision values for each tissue type, providing insight into the accuracy of the models' optimistic predictions.
The row and column summaries of the confusion matrix offered a comprehensive insight into the models' performance. We were able to examine the model's efficacy for each real class using row summaries, while column summaries revealed how effectively the model predicted each class. Heatmaps and other visual representations of the confusion matrix improved the interpretability of the data by showing the models' strengths and shortcomings in categorizing distinct tissue types.
6. RESULTS AND DISCUSSION
According to the findings, both the ResNet34 and EfficientNetB4 models perform well in classifying colorectal cancer histological tiles. However, RedNetB4 performs better in terms of total accuracy. The computational efficiency investigation shows that ResNetB4's simple and deep layer-by-layer stacking technique preserves adequate inference times.
6.1. Experiment 1: Resnet34 Results
On testing over 50 epochs at a learning rate set to 0.005, we got an accuracy of 99.976%. Confusion Matrix, Graph of ROC area obtained as shown in Figs. (6 and 7).
6.2. Experiment 2: EfficientNetB4 Results
The EfficientNetB4 has been halted at epoch 34 after 3 adjustments of the learning rate with no improvement. In this experiment, we got an accuracy of 99.886%. Confusion Matrix, Precision of dataset’s classes and graph of training and validation accuracy are generated as shown in Figs. (8-10), respectively.
 |
Fig. (8). EfficientNetB4 confusion matrix. |
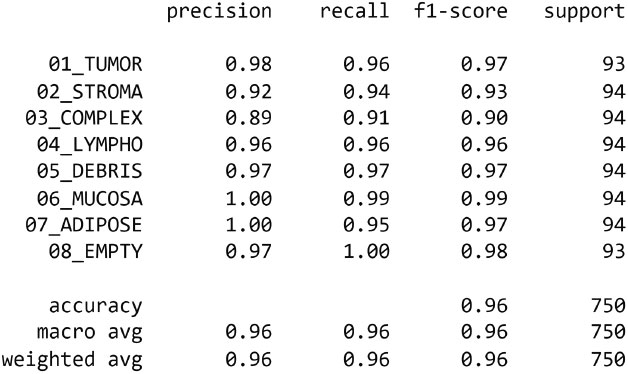 |
Fig. (9). EfficientNetB4 dataset classification results. |
 |
Fig. (10). EfficientNet34 training and validation accuracy. |
Although colorectal cancer using RESNet34 and EfficientNetB4 models have shown better outcomes, some of the points require attention. One is the potential bias introduced by the dataset used for training and evaluation, which may not accurately represent the broader population of colorectal cancer cases. This study also lacks an exhaustive analysis of other relevant architectural variants or ensemble methods, limiting the overall understanding of the deep learning landscape for colorectal cancer classification. Another limitation is the sensitivity of chosen hyperparameters to the specific dataset characteristics, which could benefit from more extensive hyperparameter tuning analysis. The lack of comprehensive model interpretability methods also hinders a deeper understanding of the features influencing the models' predictions. The paper could improve its clinical relevance by refining evaluation metrics in collaboration with medical experts. Addressing these limitations is crucial for enhancing the robustness and practical applicability of the proposed models in the context of colorectal cancer classification.
The performance of the ResNet34 and EfficientNet34 were compared with existing models and are tabulated in Table 1 and depicted in Fig. (11). ResNet34 outperforms all other models. Although these models help in the early detection of colorectal cancer, some of the well-known anti-cancer agents or treatments can be suggested to the intended audience for continuous monitoring and lifetime benefit. These include nanoparticles, which play a vital role in cancer therapy, by handling it in a patient-specific tailored way [31]. Furthermore, cancerous cases related to genetic disorders can be effectively treated using single-cell technology by examining every single cell in the tumour [32]. Also, feasible chemical complex combinations like Ruthenium and cymene can be explored for anti-cancer activity against different cells [33]. These anti-cancer activities will add great effect, if the cancer can be detected at early stages using the suggested deep learning techniques.
| Models/Refs | Epoch | Learning Rate | Accuracy |
|---|---|---|---|
| ResNet18 [17] | 30 | 0.01 | 94.16 |
| ResNet50 [17] | 30 | 0.01 | 94.85 |
| ResNet101 [17] | 30 | 0.01 | 92.74 |
| GoogLeNet [17] | 30 | 0.01 | 92.39 |
| AlexNet [17] | 30 | 0.01 | 91.92 |
| ResNet34 | 50 | 0.005 | 99.976 |
| EfficientNetB4 | 34 | 0.001 | 99.886 |
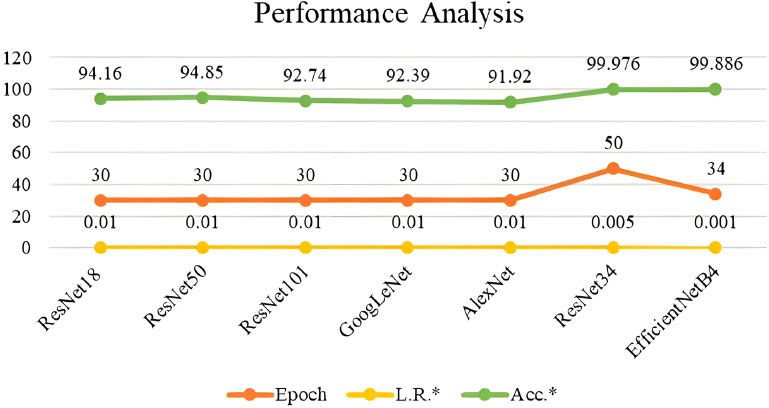 |
Fig. (11). Performance analysis of different models. |
CONCLUSION
The conclusion highlights the study's principal results and examines their consequences. New research possibilities in this arena are proposed, such as the investigation of various deep learning architectures and the inclusion of other clinical characteristics.
The key findings of the study were compared and discussed their implementation and results with existing algorithms. The research paper also highlights the limitations of the study, such as the specific dataset used and potential biases. Finally, future directions for research in this domain are suggested, including the exploration of other deep-learning architectures and the incorporation of additional clinical features. Some of the notable limitations include bias in the dataset, the sensitivity of hyperparameters remaining unexplored, and clinical analysis would further assist in evaluating the model better.
The study on colorectal cancer classification using RESNet34 and EfficientNetB4 suggests several promising future directions. It suggests exploring a wider range of deep learning architectures, including novel variants and ensemble methods, to optimize classification. Integrating transfer learning and pre-trained models on larger medical imaging datasets could improve generalization capabilities. Advanced optimization strategies, such as alternative optimizers and regularization techniques, could refine the training process. Hyperparameter tuning, possibly using automated approaches, could uncover optimal configurations for parameters like learning rates and batch sizes. Enhanced model interpretability using attention mechanisms and saliency maps is crucial for classification decisions. Real-world clinical validation, collaboration with medical professionals, and model deployment in clinical settings are essential steps toward practical application. Ensemble approaches and scalability considerations are also needed for efficient real-time deployment. Evaluating models' generalization capabilities across different medical imaging domains would provide a comprehensive understanding of their potential applications in medical image analysis.
LIST OF ABBREVIATIONS
| CNNs | = Convolutional Neural Networks |
| Adamax | = Adaptive Moment Estimation with Maximum |
| WAC | = World Automation Congress |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used in this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data and supportive information are available within the article.
FUNDING
None.
CONFLICT OF INTEREST
The author declares no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.








