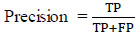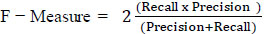All published articles of this journal are available on ScienceDirect.
A Study of Machine Learning Algorithms Performance Analysis in Disease Classification
Abstract
Background
Because there are no symptoms, it might be difficult to detect CKD in its early stages. One of the main causes of CKD is diabetes mellitus (DM), and early detection of the condition can assist individuals in obtaining prompt treatment. Because this illness has no early signs and is only discovered after the kidneys have gone through 25% damage, early-stage prediction is not very likely. This is the key factor driving the need for early CKD prediction.
Objective
The objective of the paper is to find the best-performing learning algorithms that can be used to predict chronic kidney disease (CKD) at an earlier stage.
Methods
This research aimed to compare different machine learning algorithms used in different disease predictions by various researchers. In this comparative study, machine learning algorithms like Logistic Regression, K-Nearest Neighbor, Decision Tree, Support Vector Machine, Artificial Neural Network, Random Forest, Composite Hypercube on Iterated Random Projection, Naïve Bayes, J48, Ensembling, Multi-Layer Perceptron, Deep Neural Network, Autoencoder, and Long Short-Term Memory are used in disease classification.
Results
Each classification model is well tested in a different dataset, and out of these models, RF, DNN, and NB classification techniques give better performance in Diabetes and CKD prediction.
Conclusion
The RF, DNN, and NB classification algorithms worked well and achieved 100% accuracy in predicting diseases.
1. INTRODUCTION
1.1. Overview
Due to its high mortality rate, chronic kidney disease, also known as CKD, has recently received a lot of media attention. The World Health Assembly of the WHO claims that developing nations are in danger from chronic illnesses. Unbeknownst to many, it is a disorder that frequently stays undiagnosed until the sickness is well along. The identification of CKD is often made via a physical examination of individuals who have been identified to be in danger of acquiring renal problems [1]. If CKD is detected in its early stages, it may be treated, but if it is not, itresults in renal failure. According to a study done in 2016, chronic kidney disease or chronic renal illness claimed the lives of 753 million individuals globally. Four hundred seventeen million of the 753 million individuals were women, and the other 336 million were men. This results from chronic renal disease, which affects the urine system. As the kidneys' ability to operate declines over time, they eventually fail. The waste product present in the blood also causes various health issues such as diabetes, cardiovascular disease, eye problems and blood pressure. Gloabally, nearly 10% of people were suffering from this chronic kidney disease [2]. In China, 10.8% were affected [3], and in the United States ranges from 10% to 15% were affected [4]. From another survey, 14.7% of people were affected in the Mexican population [5]. The total loss of renal function is the primary symptom of this CKD condition. In its early stages, this disease will not show any of the symptoms; hence, it is identified after we lose kidney function by 25% [6]. They exhibit acute malfunction in immunological and neural disorders, which have detrimental effects on patients who are affected in their daily routines by chronic kidney disease (CKD). Kidney failure is brought on by the major problem in CKD. It leads to kidney transplant or else it leads to end-stage life span [7]. Therefore, by providing appropriate therapy and being able to anticipate CKD, the effects may be minimized. Here are a number of diagnostics and computing methods utilized in both the detection and evaluation of CKD complexity [8]. People from developed countries were not aware of this disease, which led to dialysis or transplants. To measure CKD, we have a variety of approaches, including computational ones. By assessing the rate at which glomerular filtration occurs (GFR) based on the patient's age, blood type, sex, cholesterol levels, weight, and other characteristics, this condition was discovered. The GFR value is classified into five stages [9]. Table 1 shows different GFR levels.
| Stages | GFR (mL/min/ 1.73 m2) | Description |
|---|---|---|
| I | ≥90 | Kidney Function Fair |
| II | 60–89 | Slight CKD |
| III | 30–59 | A Medium CKD |
| IV | 15–29 | Severe CKD |
| V | ≤ 15 | End Stage CKD |
To measure the GRF, we need two test values: one is from the blood test to check creatinine, and another is the urine test to check albumin [10]. This traditional method is used to identify the presence of albumin and creatinine, which calculates the GFR and the levels of kidney functions.
One of the main causes of chronic kidney disease (CKD) is Diabetes Mellitus (DM), and those with the condition are more likely to develop renal failure. People affected by diabetes have a lifelong disorder; taking regular treatment and prevention is very difficult; the medical expenses are also high, and the price is not affordable for average and below-average people. Regular monitoring and giving treatment are also tough for doctors. Because the pancreas does not create enough insulin, this kind of diabetes develops. Diabetes is mainly classified into three categories: gestational diabetes (Diabetes during pregnancy), type 1 (T1DM), and type 2 diabetes (T2DM). Insulin secretion by the pancreas fails in type 1 diabetes [11], hence the need of taking insulin physically. Type 2 diabetes occurs when not enough insulin is secreted from the pancreas to filter proteins in the blood. Gestational diabetes occurs during pregnancy due to an excess intake of food that causes insufficient insulin to process the blood. In this modern culture, CKD affects most people around the globe, and Type 2 diabetes is also one of the root causes of CKD. Hence, reducing the disease, monitoring it, and finding it is most important here. Patients with nephropathy or heart problems might have increased GFR values; such patients missed out on this CKD criteria. Furthermore, there is not enough testing and monitoring equipment, as well as inadequate doctors to observe every patient with CKD every time. The computer-based monitoring system needed for CKD diagnosis and testing is economical and affordable. Artificial intelligence techniques are used in most areas of the field for early prediction [12]. In the medical field, for disease prediction and classification, an advanced technique called machine learning is very useful for earlier detection. The data preprocessing and classification are done using Machine learning.
The intention of investigating Chronic Kidney Disease, also referred to as CKD, is to find the medical condition at an early stage and stop it from developing into end-stage renal failure, which is severe in the absence of kidney transplantation or external filtration (kidney function). Being able to prevent a medical condition's development via prompt care makes the diagnosis of chronic renal failure (CKD) crucial. The likelihood of a pause in the development of the illness increases with early detection. A correct diagnosis also makes it possible to start taking medications and altering the way one lives in order to control the illness and stop the consequences. Furthermore, knowing the underlying factors underlying kidney failure may aid in creating plans aimed at preventing the illness from ever starting in its preliminary region.
1.2. Paper Contribution
The next few reasons encapsulate all primary motives behind this work.
- In order to choose the best characteristics with regards to machine learning, researchers investigate several methods for selecting features.
- We aim to identify the most suitable feature group that has been verified clinically and also chosen from a machine learning standpoint.
- Collaborating through a healthcare professional, we identify the highest impacted aspects through the clinician's viewpoint as we examine the clinical diagnosis of chronic kidney disease.
- Using several subgroups of features from the chronic kidney disease set of data, researchers develop and analyse base-learning models.
- Our objective is to evaluate each system's efficiency in comparison to comparable variants.
1.3. Literature Review
The state of the art of CKD evaluation and prediction is addressed in this section. Our major areas of interest are feature-selection techniques and Machine Learning models with Deep Learning. Most studies focus on previous and laboratory test records to build promising predictive models. For example, Gazi Mohammed Ifraz et al. [13] employed machine learning techniques to classify and predict chronic kidney disease from the CKD dataset available in Kaggle. From this approach, the Logistic Regression (LR) performs well and produces an accuracy of 97%. Another approach by Hashi et al. [14] for the presence and absence of CKD prediction was done through the K-Nearest Neighbor (KNN) classification algorithm with 76.96% accuracy. In the study from Alasker et al. [15], which uses a Decision Tree (DT) classifier for predicting kidney disease, this Decision Tree (DT) results in 98.4127% accuracy by using all 24 features from the dataset taken from the UCI ML repository. Baitharu and Pani [16] employed an artificial neural network (ANN) for classifying the disease in the healthcare system using a dataset of liver disease, which produced an accuracy of 71.59%. In their study, Bashir et al. [17] developed a disease prediction model by using a multilayer classifier. They used K-Nearest Neighbour (KNN) for early disease prediction, which achieved 57.41% accuracy from the Statlong dataset. Khan et al. [18] utilized a KNN and Random Forest (RF) classifier to predict liver disease from the dataset taken from the UCI ML repository, KNN produced an accuracy of 62.90% and RF of 72.17% accuracy. An approach from Vijayarani et al. [19] employed an ANN classification to predict kidney disease in a dataset taken from different health centres like hospitals, labs, and medical centres. This ANN classification achieves an accuracy of 87.70%. Another approach by Dar and Azmeen [20] uses this decision tree (DT) classification technique for predicting dengue fever based on real data collected from various hospitals. This DT classification produced an accuracy of 76%. An approach from Pahareeya et al. [21] proposed a classification model for liver disease prediction from the liver dataset in the UCI ML repository. This SVM achieves an accuracy of 71.5026%. Khan et al. [22] employed a Composite Hypercube on Integrated Random Projection (CHIRP) for classifying kidney disease as positive or negative, using the dataset taken from the UCI ML repository. This CHIRP produces an accuracy of 99.75%. Rashed-Al-Mahfuz et al. [23] use the Random Forest (RF) classifier to predict the CKD or NOTCKD from the CKD dataset, which was taken from the UCI ML repository. RF achieves an accuracy of 99.50% for the selected features. Jeong et al. [24] used the Autoencoder (AE) for classifying the chronic kidney disease stages with a highly imbalanced dataset taken from the National Health Insurance Corporation (NHIC) in Korea. This AE achieves an accuracy of 99.58%. Rahman et al. [25] developed a diabetes disease prediction classification model using Conv-LSTM from PIDD in the National Institute of Diabetes and Digestive Diseases. The Conv-LSTM-based technique performs well and achieves 97.26% accuracy. Garca-Ordás et al. [26] proposed a new approach based on the combination of a Sparse Autoencoder and a Convolutional classifier in diabetes classification from the Pima Indians Diabetes Dataset (PIDD). This model achieves an accuracy of 92.31% in disease classification. Nadesh et al. [27] developed a model to predict Type 2 diabetes mellitus from feature selection and Deep Neural networks. This proposed model performs well and achieves 98.16% accuracy using the Pima Indians Diabetes Dataset (PIDD) taken from the UCI ML repository. Senan et al. [28] used various ML classification techniques for CKD disease prediction like Random Forest, K-Nearest Neighbors, Decision Tree, and Support Vector Machine. The CKD dataset of 400 patients was collected from the UCI ML repository. This approach gives the best diagnostic results in disease classification and produces accuracy ranges of SVM 96.67%, KNN 98.33%, DT 99.17, and RF 100%. Sunge et al. [29] developed a diabetes prediction model using different Machine learning techniques, including Decision Tree (DT), K-Nearest Neighbors (KNN), Naïve Bayes (NB), and Artificial Neural Network (ANN) datasets taken from the Pima Indians Diabetes Dataset (PIDD). From this model, the KNN model achieves an accuracy of 80.34% in diabetes prediction. Another approach by Ranjith et al. [30] proposed a model to predict diabetes disease using various ML algorithms. The algorithms are K-Nearest Neighbors (KNN), Decision Tree (DT), Naïve Bayes (NB), Deep Neural Network (DNN), RL, and LR. With this algorithm, DNN performs well and produces 100% accuracy. Meng et al. [31] developed various ML models to predict pre-diabetes. These classification algorithms (LR, DT, and ANNs) are used to analyse risk factors and predict pre-diabetes. DT performs well and produces 77.87%. Aljaaf et al. [32] used the MLP neural network (MLP) to predict early-stage CKD; this approach achieves 98.1% accuracy. Subasi et al. [33] proposed a CKD diagnosis model using various Machine Learning (ML) classification techniques. The SVM, MLP, C4.5 DT, KNN, and RF were used, and Random Forest outperformed them well and achieved 100%. Another approach by Boukenze et al. [34] used MLP to predict Chronic Kidney failure; this model produces 99.75% accuracy. Almansour et al. [35] employed Neural networks (NN) and Support Vector machines (SVM) to predict CKD disease; this approach performed well, and both produced 97.75% accuracy. Gunarathne et al. [36] used a Decision Tree (DT) to predict CDK presence; this classification model achieves 99.1% accuracy. Another study by Kunwar et al. [37] uses two Machine learning (ML) techniques: Naïve Bayes (NB) and Artificial Neural Network (ANN) for CKD disease classification datasets taken from the UCI ML repository. Naïve Bayes performs well and produces an accuracy of 100%. Avci et al. [38] developed a disease classification model to predict CKD presence and absence. This model uses J48, SVM, NB, and K-Star to diagnose the UCI ML dataset for disease prediction. In this model, the J48 classifier achieves 99%. The study from Aliberti et al. [39] used NAR and an LSTM model for blood glucose prediction; this LSTM produces 88.55% accuracy in the not filtered training set and 99.73% accuracy in the filtered training set. The RT_CGM dataset was taken from SAFHS. Pradhan et al. [40] employed Artificial Neural Network (ANN) for diabetes prediction. This model predicts diabetes with an accuracy of 85.09% based on a dataset taken from the Pima Indian Diabetes Dataset (PIDD). Islam et al. [41] used various machine learning (ML) techniques, but the ensembling approach outperformed the others in predicting Type 2 diabetes. This model produced 95.94% accuracy in the disease classification model. The study by Dritsas et al. [42] developed a CKD prediction model using the Random Forest (RF) classification algorithm. This RF diagnosis the CKD dataset from the UCI ML repository to predict CKD with an accuracy of 99.2% [43]. This paper presents an adaptive interference removal framework (IRF) for video person re-identification (V-ReID) to improve accuracy [44]. A novel feature learning framework is proposed to capture significant information in spatial and temporal domains, building a discriminative and robust feature representation for each sequence [45]. This study investigated the associations of behavioral and health-related factors with chronic kidney disease (CKD) in Iranian patients. A hospital-based case-control study found that low birth weight, diabetes history, kidney disease history, and chemotherapy history are associated with CKD risk. The results highlighted the importance of collaborative monitoring of kidney function among patients with these conditions [46]. To solve the shortcomings in current picture paint techniques, a suggested restored image consider blends linguistic assumptions with a thorough resultant group. It comprises of a Natural language Priors Network, which is Deep Attention Residual Company, and Full-scale Remove Connection. The approach concentrates on stream features learning logical prior knowledge for deficient areas, and repairs defective regions via full-scale jump joins. Current state-of-the-art approaches are surpassed by the method [47]. This study suggests a lightweight, single-image super-resolution system that combines multi-level characteristics to solve typical issues such as slowly convergent images and blurry edges. The suggested approach works much better when factors are large, and it operates present methods by means of individual viewpoints and independent metrics [48]. The study suggests a two-stage picture inpainting network that is superior and depends on situational perception and similar networks. Research on public datasets show that the technique delivers an increased likelihood of a mending outcome [49]. The study suggests a stochastic adversarial network-based technique for efficient picture inpainting. The combination of these modules enhances the visual impact and quality of the images, surpassing current standards in the two types of assessments [50]. In order, the level of imagine inpainting increases. This research suggests a simple method that employs grouping combination along with the attention mechanism. Comparing experimental findings with similar light-weight strategies, infer less time and used resources are observed.
2. MATERIALS AND METHODS
In the following phase, we will deeply discuss various algorithms in Machine learning that are employed to segregate the diseases. We have a large number of algorithms for designing the classification model. Here, we will see some of the classification techniques which are used. The algorithms are Logistic Regression (LR), K-Nearest Neighbour (KNN), Decision Tree (DT), Support Vector Machine (SVM) Artificial Neural Network (ANN), Random Forest (RF), Composite Hypercube on Iterated Random Projection (CHIRP), Naïve Bayes (NB), J48, Ensembling, Multi-Layer Perceptron (MLP), Deep Neural Network (DNN), Autoencoder (AE), and Long Short-Term Memory (LSTM) used in disease classification.
2.1. Logistic Regression (LR)
Logistic regression is one of the accurate classification models used in the medical industry. For independent characteristics, the LR, which is usually used to forecast the class variable, produces a probability output of 0 or 1.
2.2. K-Nearest Neighbour (KNN)
K-Nearest Neighbour is the simplest method to classify the labels from the given dataset. The distance between the unlabelled instances in the class was measured with the nearest instance present in the class. It uses the k-Nearest Neighbours classifier.
2.3. Decision Tree (DT)
A decision tree is the most popular method used to classify the data; based on the decision, the tree will grow gradually. It is used for all kinds of real-time problems to make the right decision. DT has a higher number of layers, which reduces the performance and is also a little more complex. This has an overfitting problem in data handling.
2.4. Support Vector Machine (SVM)
Support Without using the probability approach, vector machines are a classification technique used to determine the class labels from the dataset. By using the same reasoning to get the class labels for linear equations. There will be two approaches to problem-solving: linearly separable and non-linearly separable.
2.5. Artificial Neural Network (ANN)
The fundamental function of neural networks is the use of neurons to carry out communication between the brain and other areas of the human body. Using artificial neural networks, we may employ this logic to convey information and carry out various actions.
2.6. Random Forest (RF)
A random forest is created by the combination of several Decision trees. The appropriate Subset is classified from the dataset through decision-making. The output from the several decision trees should be categorised based on the majority vote.
2.7. Composite Hypercube on Iterated Random Projection (CHIRP)
Using CHIRP, the prediction has the highest accuracy compared to normal prediction. For 2D prediction and symmetrical location prediction, it requires analysing a single set of data; the analytically effective methods are used by CHIRP. All predictions are grouped together by the CHIRP classifier and made into new data.
2.8. Naïve Bayes (NB)
Compared to numeric data, the Naive Bayes Classifier performs better, however it can only be used with a certain kind of dataset. One of the several categorization methods used in Bayesian learning is the naive Bayes algorithm. The Naive Bayes Classifier and Bayesian Belief Networks are two common algorithms that use Bayesian learning.
2.9. J48
Data collection in J48 is carried out via a divide-and-conquer top-down recursive strategy.
2.10. Ensembling
In Machine learning, ensembling is used to predict more accurately than a single classification model. This method will combine the different individual models for better performance in output prediction. Two methods were used here: stacking and voting. Soft approaches are used in voting in order to get a high probability, and the Staking procedure then yields the final forecast.
2.11. Multi-Layer Perceptron (MLP)
With a multi-layer perceptron, we may add additional layers to the model to forecast better outcomes. For greater performance in the additional layers, we apply a variety of activation functions. Step function, soft maximum function, sigmoid function, and more. All updated weights are to be done through the backpropagation approach in MLP.
2.12. Deep Neural Network (DNN)
Along with a Deep Neural Network (DNN), an artificial neural network, the input and output include numerous hidden models. This is the main characteristic of the Neural network: it processes complex progressive data in real-time. The forward propagation method is used for data processing from input to output.
2.13. Autoencoder (AE)
It is a data compression technique that is used for compressing the data in the hidden layer of a neural network. The encoder is used to decrease the hidden units in the input layer, and the decoder is used to increase the hidden units in the output layer. The error handling is done through the backpropagation method.
2.14. Long Short-Term Memory (LSTM)
Long Short-Term Memory is used to store and process data for a long time. To reduce the vanishing gradient problem, the LSTM was used. It has some memory space for temporary storage. LSTMs employ a Tanh layer and three logistic sigmoid gates to control memory. The output for this model is the binary numbers 0 or 1.
2.15. Proposed Method
With the purpose of comparing many cutting-edge machine learning techniques for CKD detection, approaches for selecting features served for which traits were best chosen. Medically, CKD is impacted by the selected characteristics. The chosen dataset, the preprocessing stage, the feature-selection techniques, the ML model, and the optimisation techniques comprise the whole suggested approach's stages. The outcomes might change based on each suggested machine learning model.
3. PERFORMANCE EVALUATION METRICS
3.1. Confusion Matrix
Table 2 shows the performance evaluation of the model.
Using the evaluation metrics, we evaluate the model to validate the performance. The metrics are Accuracy, Recall, Precision and F- measure.
| Class | 1(Positive) | 0(Negative) |
|---|---|---|
| 1(Positive) | TP-True Positive | FP-False Positive |
| 0(Negative) | FN-False Negative | TN-True Negative |
3.2. Accuracy
When more than half of a classification's predictions come true, it is said that the expected value and the actual value are equal.
 |
(1) |
3.3. Recall
Recall is used to calculate the system's ability to predict the number of positive samples
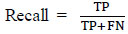 |
(2) |
4. RESULTS AND DISCUSSION
In this section we discuss the algorithm performance in the disease prediction from various aspects.
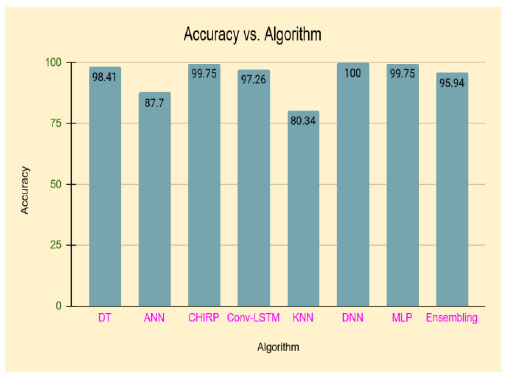
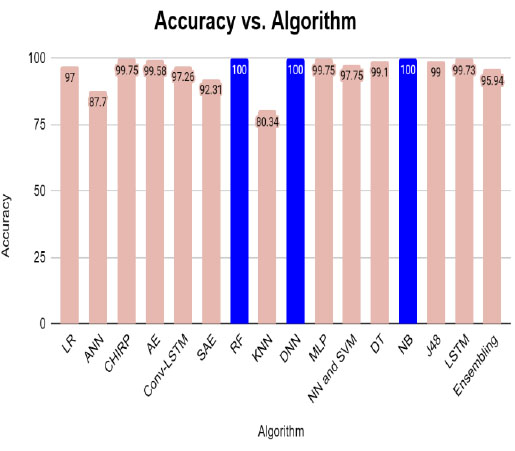
Table 3 represents the different algorithms used for disease prediction with different accuracy ranges in various diabetes datasets. This comparison shows that the DNN outperforms well in the diabetes dataset and produces higher accuracy than the remaining algorithms. Fig. (1) shows the pictorial representation of comparison results.
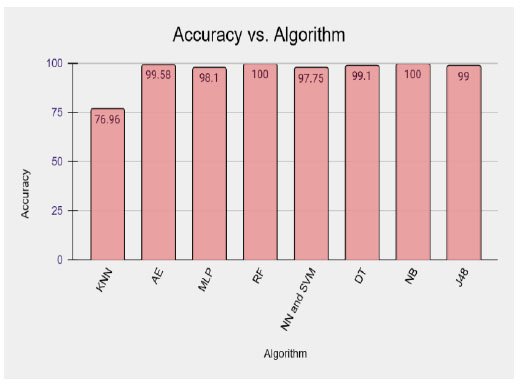
Table 4 represents the different algorithms used for disease prediction with different accuracy ranges in various CKD datasets. This comparison shows that the RF and NB outperformed well in the CKD dataset and produced higher accuracy than the remaining algorithms. Fig. (2) shows the graphical representation of the results.
Table 5 represents the various algorithms used for disease prediction with different accuracy ranges in both the diabetes and CKD datasets. This comparison result shows that the DNN, RF, and NB outperformed well in both datasets and produced higher accuracy than the remaining algorithms. The graphical representation shown in Fig. (3).
| Algorithm | Accuracy (%) |
|---|---|
| LR | 97 |
| ANN | 87.7 |
| CHIRP | 99.75 |
| AE | 99.58 |
| Conv-LSTM | 97.26 |
| SAE | 92.31 |
| RF | 100 |
| KNN | 80.34 |
| DNN | 100 |
| MLP | 99.75 |
| NN and SVM | 97.75 |
| DT | 99.1 |
| NB | 100 |
| J48 | 99 |
| LSTM | 99.73 |
| Ensembling | 95.94 |
CONCLUSION
The comparative study observed that Machine Learning techniques will help to change the scenario in the medical sector. In this paper, we compared various algorithms for predicting the disease with different precision ranges from various studies. Finally, as observed from Table 5, DNN, RF, and NB outperform well with an accuracy of 100%. This comparison study clearly highlights that DNN, RF, and NB are the most promising techniques for Disease prophecy.
Although the approach we recommend draws upon aspects that are shown to make a major impact on CKD classification, it makes sense from the standpoint of medicine. We want to make sure that our suggested methodology can be used broadly by expanding our research later on by obtaining real-world data in academic hospitals. In addition, since long-term kidney problems may coexist having various medical conditions, they are interested in exploring the relationship between the state of all medical conditions with the overall wellness of people having chronic renal failure. Finally, we want to investigate the produced technique's rate of computation.
LIST OF ABBREVIATIONS
| DM | = Diabetes Mellitus |
| CKD | = Chronic Kidney Disease |
| RF | = Random Forest |
| NB | = Naïve Bayes |
| DNN | = Deep Neural Network |
| WHO | = World Health Organization |
HUMAN AND ANIMAL RIGHTS
No humans/animals were used for studies that are the basis of this research.
AVAILABILITY OF DATA AND MATERIALS
The data supporting the findings of the article is available in the UCI ML repository, Kaggle and Pubmed at URL [https://archive.ics.uci.edu/dataset/336/chronic+kidney+disease, https://archive.ics.uci.edu/dataset/857/risk+factor+prediction+of+chronic+kidney+disease, https://pubmed.ncbi.nlm.nih.gov/22386035/, https://www.kaggle.com/datasets/uciml/pima-indians-diabetes-database diabetes.csv].