All published articles of this journal are available on ScienceDirect.
Multi-class Classification of Gastrointestinal Diseases using Deep Learning Techniques
Abstract
Objective:
Gastrointestinal problems have claimed the lives of about two million individuals worldwide. Video endoscopy is one of the most important tools for detecting gastrointestinal diseases (GD). However, it has the disadvantage of producing a huge quantity of images that should be analysed by medical professionals, which demands more time. This makes manual diagnosis challenging, encouraging research on computer-assisted methods for accurately identifying all of the produced images in a short amount of time. The goal of this research is to develop an artificial intelligence technique to classify different gastrointestinal problems.
Materials and Methods:
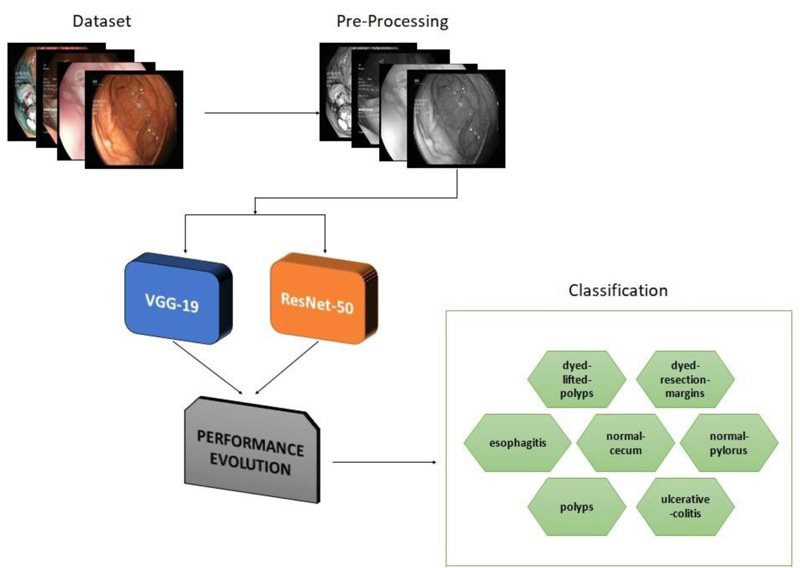
In developing a framework for classifying gastrointestinal diseases, the proposed methodology is unique. VGG-19 and ResNet-50, two deep learning networks, were used to evaluate their ability to recognise a dataset of lower GD. To validate the proposed models, a Kvasir dataset of 3500 images was employed, comprising 500 images from each of the seven classes.
Results:
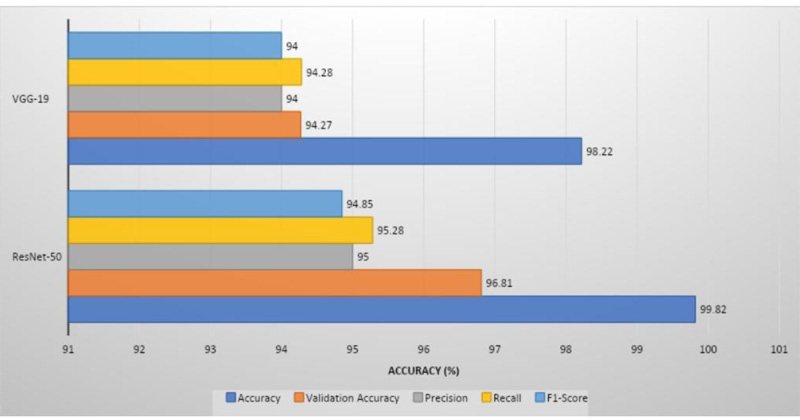
ResNet-50 had the greatest validation accuracy of 96.81%, recall of 95.28%, precision of 95%, and 94.85% of F1 score, whereas VGG-19 had the highest validation accuracy of 94.21%, recall of 94%, precision of 94.28%, and 94% of F1 score.
Conclusion:
The Convolutional Neural Network (CNN) was used to extract shape, colour, and texture features. Softmax assists in the classification of seven types of GD by passing these features into fully connected layers. Two models performed equally well and yielded promising results.
1. INTRODUCTION
In comparison to other malignancies, gastrointestinal cancer is the most prevalent disease. A total of 1.8 million people die each year from gastrointestinal disorders, according to the World Health Organization (WHO) [1]. Gastroscopy tools are not appropriate for detecting and examining GD infections, such as polyps, ulcers, and bleeding, because of their intricacy. Wireless capsule endoscopy was developed in 2000 to tackle the difficulty with gastroscopy equipment [2]. According to the 2018 annual report, the above equipment was used on almost one million patients [3].
GD infections, particularly in the small intestine, can be identified, but the overall method takes a long time to evaluate the video frames obtained from each patient. On the other hand, physicians face difficulties that need an excess of time to evaluate all of the data [4]. In various medical fields, artificial intelligence (AI) technologies have shown great potential in aiding humans in visualising disorders that are not evident to the naked eye. The application of two CNN models for the identification and classification of gastrointestinal disorders is the subject of this article. Machine learning is highly useful in medical diagnoses, including lung disease, chest, etc. [20-22].
By incorporating engineering into learning, CNN has demonstrated its remarkable ability to abstract features. In medical image diagnostics, deep learning algorithms have proved that they can outperform specialists [5]. Consequently, computer-assisted diagnoses for endoscopic images based on deep learning algorithms are projected to attain diagnostic accuracy comparable to that of qualified specialists [6]. Igarashi et al. presented the AlexNet model for classifying upper gastrointestinal organs, which had a 96.5% accuracy [7].
Biniaz et al. employed factorization analysis, a technique for speeding up endoscopic screening evaluation [8]. The sliding window mechanism with singular value decomposition was also utilized, which offered an overall accuracy of 92%. Cogan et al. employed a variety of CNN pre-trained models, including Inception V2, Inception V4, NasNet, and ResNet-V2 [9]. With Inception V4, a classification accuracy of 93.8% can be achieved. AbraAyidzoe et al. suggested a Gabor capsule network that classifies the Kvasir dataset images with a 93.65% accuracy [10]. Song et al. employed CNN to classify polyps [11]. Mohaputra et al. employed the Wavelet transform in conjunction with the CNN model to reach a 93.65% accuracy [12]. To extract information from the image caption and also in the text, a sequential approach and a multimodal one were proposed by Aggarwal et al. [13] (Table 1).
The rest of the article is organized as follows: the materials and methodologies are discussed in Section 2. The simulation results are discussed in Section 3. The conclusions are highlighted finally.
2. MATERIALS AND METHODOLOGIES
Computer-assisted automated detection of GD is becoming a hot topic of research. In this section, VGG- 19 and ResNet-50 are described in detail. Fig. (1) depicts a generic block diagram for early diagnosis of GD. To extract features from the image, CNN is employed. Diagnosis and classification are aided by the fully connected layers [20-22].
2.1. VGG-19 Model
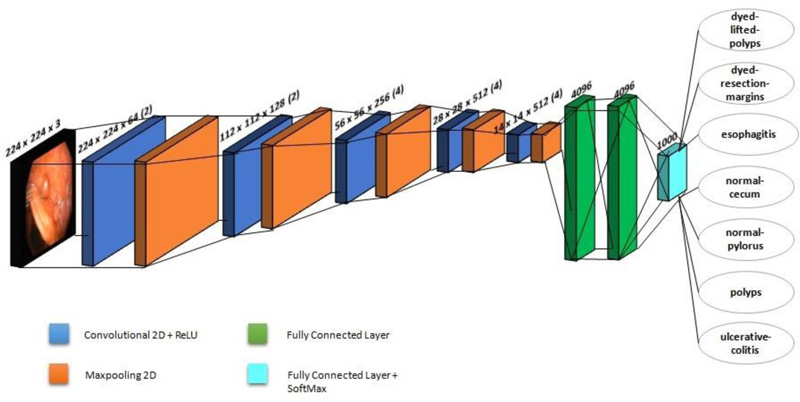
VGG-19 is a CNN with 19 layers trained on over a million images in the ImageNet database. This model has six core structures, each of which is composed of various connected convolutional layers and fully-connected layers. Fig. (2) shows the architecture of VGG-19, which is employed in this investigation. A fixed image size of 224 x 224 RGB is used in the architecture. It makes use of a 3 x 3 kernel. Spatial paddings are employed before 2 x 2 max pooling to preserve the spatial resolution. Rectified linear unit (ReLu) is used to increase classification while reducing computing time. This model is employed as a pre-processing model, and it has better network depth than the classic CNN. Multiple convolutional layers and activation layers with non-linear functionality provide a superior and alternating structure. The layer structure may abstract image features, then downsample them using max-pooling and alter the ReLU to indicate the major value in the image region as the area's pooled value. The downsampling layer increases the anti-distortion capability of the network. By considering only the important features, it reduces the parameters. An Adam optimizer is also utilised in this study, with a learning rate of 0.0001.
2.2. ResNet-50 Architecture
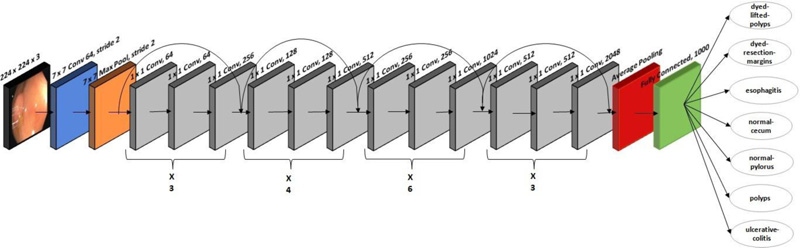
A residual CNN model with 177 layers is used in ResNet-50 architecture. ResNet-50 won the image classification competition in 2015. The ResNet-50 network is mainly used in computer vision applications. The architecture of ResNet-50 is divided into four stages. The network can accept an image with a height and width that are multiples of 32 and a channel width of 3; 224 x 224 x 3 is the input size specified in the design. Every ResNet architecture adopts a kernel size of 7 x 7 for initial convolution and 3 x 3 for max pooling. Stage 1 contains three residual blocks, each with three levels. The kernels used to execute convolution operations in all three levels of stage 1's block are 64, 64, and 128. The channel width doubles, and the input size becomes half as we progress from one stage to the next. The network contains an average pooling layer, especially towards the end, succeeded by a fully connected layer of 1000 neurons. Fig. (3) shows the ResNet-50's architecture, which is used to classify 3500 images into seven lower digestive tract disorders. It includes 16 input layers and 177 layers between them. After that, only positive data is then transmitted via the convolutional layers. In this study, an stochastic gradient descent (SGD) optimizer with a learning rate of 0.00001 is employed.
2.3. Dataset under Study
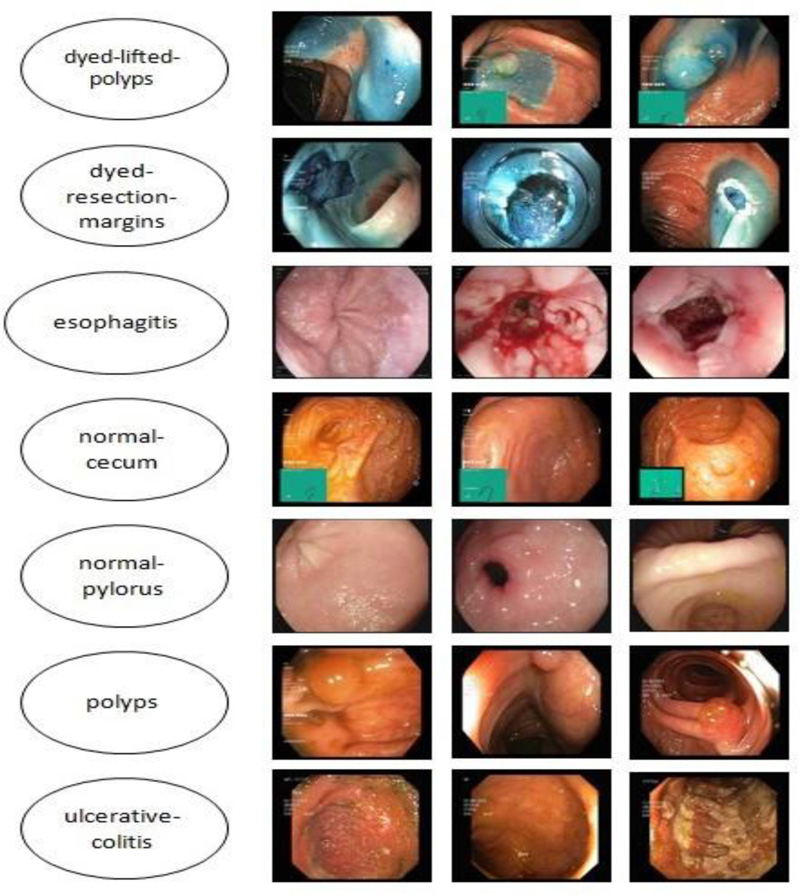
The Kvasir dataset, which comprises images of the gastrointestinal system acquired with endoscopic equipment, was used in this investigation. Fig. (4) shows sample images with subclasses from the proposed study, which includes seven classes with 500 images each. The images are resized to 224 × 224 pixels.
2.4. Preprocessing and Augmentation Techniques
Due to the increased complexity of feature extraction, noise and artifacts caused by light reflections, camera angles, and the mucosal membranes covering internal organs degrade CNN performance. Therefore, researchers have been interested in optimization approaches to enhance image quality. After resizing the image to 224 × 224 pixels, an average filter is used as the final step in the improvement process. It replaces each pixel by averaging it with its neighbours, a procedure that is repeated for all the pixels in the image.
2.5. Convolution Layers
Numerous features, including shape, texture, and colour, are present in the gastrointestinal dataset. When extracting images from a video, many images do not include the disease, and the disease features only exist in a few images that may be overlooked by the radiologist and the specialists. Hence, manual extraction of features requires significant skill. However, in the case of CNN algorithms, convolutional layers are used to extract representative aspects of each disease. VGG-19 is a CNN with 19 layers, and the ResNet50 CNN model has 177 layers. The classification layers have the 9216 representative features that the convolutional layers extract from each image and represent as feature maps.
2.6. Normalization and Dropout Technology
In order to speed up training procedures for deep neural networks, normalisation is one of the approaches utilised during training. Gradient descent converges normalisation and assists in selecting the proper learning rate. The learning rate is more challenging and requires more time without the normalisation process. In our research, the mean of the entire training set for each pixel was subtracted during the image normalisation process. Data was centered, and each feature's variance was set to one by computing the dataset's variance and dividing it by each pixel. Millions of parameters are generated by CNNs, which causes overfitting. In order to lessen overfitting, CNN uses a dropout method. In our research, the dropout approach was used to stop 50% of the neurons in ResNet-50 and 40% for VGG 19 in each iteration. As a result, each iteration of the network used a distinct set of parameters. Hence, the training time doubled with the dropout technique.
2.7. Transfer Learning and Optimizer
Transfer learning is based on using a dataset to train on a particular problem and then applying that knowledge to another problem with a different dataset. In order to apply what has been learned to a different task, transfer learning selects the pre-trained model and the size of the challenge. Overfitting is also avoided through transfer learning. In this study, the weights of ResNet-50 and VGG 19 were adjusted using transfer learning. The gastrointestinal dataset was used to apply the learning from the ImageNet dataset to the ResNet-50 and VGG 19 models. Optimizers were primarily utilized to tune up the weights, bias, and learning rate of a model. The Adam optimizer was used by AlexNet, whereas the (SGD) optimizer was used by ResNet- 50.
3. RESULTS AND DISCUSSION
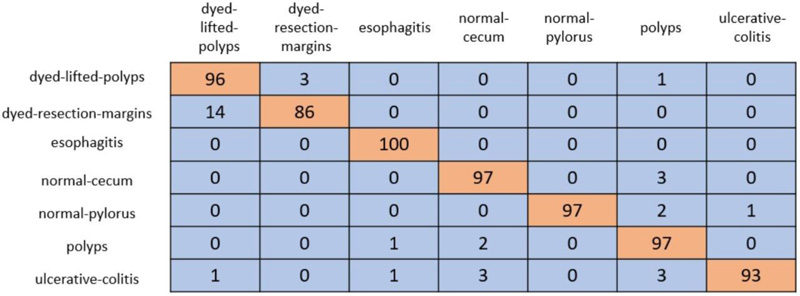
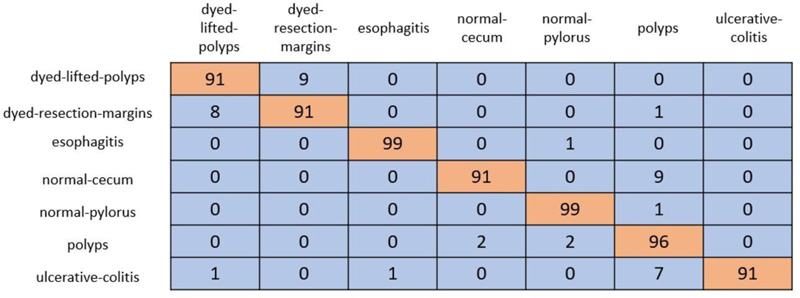
In cross-validation, 80% of the dataset was used for training, and the remaining 20% was used for testing. A coLab Pro environment is used to simulate the whole model. The confusion matrix produced from ResNet-50 and VGG-19 is shown in Figs. (5 and 6).
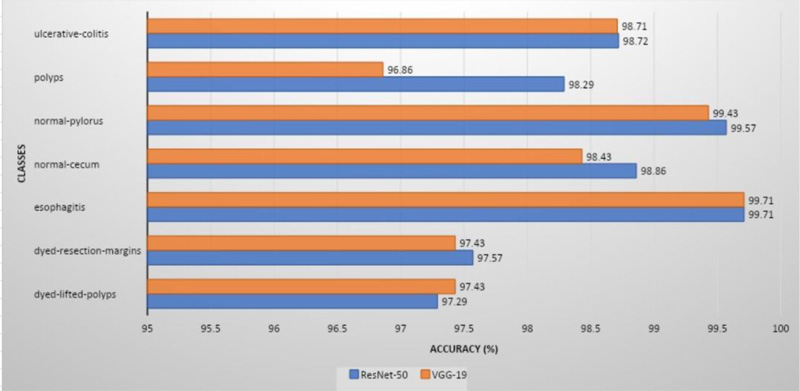
ResNet-50 showed the greatest validation accuracy of 96.81%, whereas VGG-19 had the highest validation accuracy of 94.21%. Fig. (7) depicts the models' overall metrics, whereas Fig. (8) depicts the metrics derived for each disease.
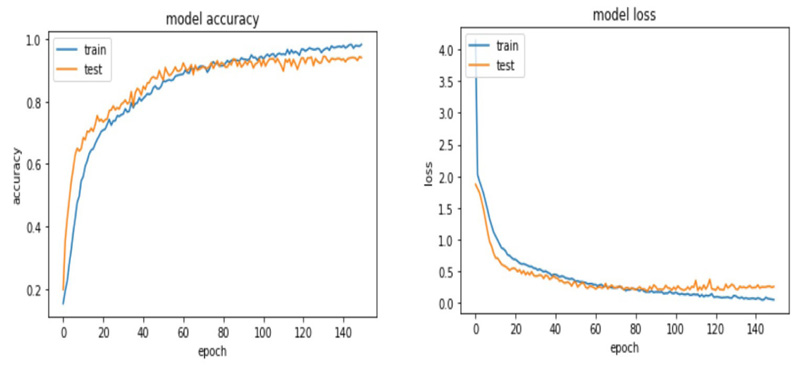
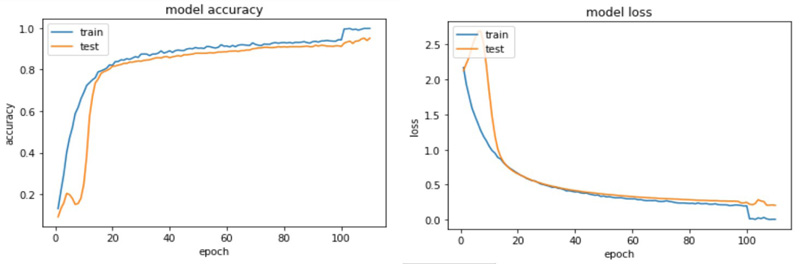
The accuracy and loss plots for VGG 19 and ResNet 50 are shown in (Figs. 9 and 10).
The proposed models were compared to models already published in the literature, as shown in Table 1. ResNet-50 was reported to have a validation accuracy of 99.82%, which was higher than the other models. Fig. (9) displays a graphical representation of performance metrics of models’ understudy compared to other models.
| Related Studies | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|
| Godkhindi and Gowda [14] | 88.56 | 88.77 | 87.35 |
| Pozdeev et al. [15] | 88 | 93 | 82 |
| Bour et al. [16] | 87.1 | 87.1 | 93 |
| Min et al. [17] | 78.4 | 83.3 | 70.1 |
| Fonoll´a et al. [18] | 90.2 | 90.1 | 90.3 |
| Mosleh et al. – AlexNet [19] | 97 | 96.8 | 99.2 |
| Mosleh et al. –ResNet-50 [19] | 95 | 94.8 | 98.8 |
|
ResNet-50 (Model understudy) |
99.82 | 95.23 | 99.16 |
|
VGG-19 (Model understudy) |
98.22 | 94.3 | 99 |
CONCLUSION
Using the Kvasir dataset, this research proposes a robust method for the classification of gastrointestinal disorders. Deep learning algorithms can help to lessen the chance of emerging cancer by assisting in early detection and reducing the need for unnecessary begin tumour excision. Despite the fact that video endoscopy is the most prevalent investigative approach for finding gastrointestinal polyps, a number of human aspects contribute to inaccurate gastrointestinal illness diagnosis. VGG-19 and ResNet-50, two deep-learning models studied in this study, can direct doctors’ attention to the vital areas that may have been neglected previously. A total of 9216 characteristics were retrieved and sent to the 1000 neurons in fully connected layers. Each image was categorised into one of the seven categories of gastrointestinal disorders by the softmax layer, which generated seven classes. The VGG 19 and ResNet50 models produced results that were quite promising. Therefore, future applications of advanced deep learning algorithms are suggested. However, the algorithm's weakest element is the need for manual data marking. As accurate disease diagnosis is frequently challenging for even humans, the network may therefore inherit certain errors from an analyst. One method to overcome this restriction is to use a larger dataset that has been labelled by a broader group of experts.
LIST OF ABBREVIATIONS
| GD | = Gastrointestinal Diseases |
| CNN | = Convolutional Neural Network |
| WHO | = World Health Organization |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used in the studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data supporting the findings of the article is available in the Kaggle at https://www.kaggle.com/datasets/plhalvorsen/kvasir-v2-a-gastrointestinal-tract-dataset.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflicts of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.


