Detection and Classification of the Different Stages of Alzheimer’s Disease using Sequential Convolutional Neural Network
Abstract
Background:
Alzheimer's disease (AD) is a degenerative disorder known as mild or significant neurocognitive impairment diagnosed in older people and significantly reduces their quality of life. Memory issues are regularly one of the principal indications of Alzheimer's disease; however, beginning manifestations may vary from one individual to another. Detecting Alzheimer's disease is a time-consuming and challenging task but requires brain imaging reports and human expertise.
Objective:
More accurate and early diagnosis is essential for timely treatment and risk reduction. The accurate diagnosis of Alzheimer's disease plays an essential role in inpatient treatment, especially at the disease's early stages, because risk awareness allows the patients to undergo preventive measures even before the occurrence of irreversible brain damage. The main objective of this research is to diagnose Alzheimer's disease in its early stage through Magnetic Resonance Imaging (MRI).
Methodology:
To detect and classify the various stages of Alzheimer’s disease from the MRI, a Sequential Convolutional Neural Network (S-CNN) model is proposed, which incorporates the advantage of convolution operation and recurrent operation.
Results:
The combination of Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset and the Open Access Series of Imaging Studies (OASIS) dataset obtained from the Kaggle is used for training and testing the proposed S-CNN model.
Conclusion:
The baseline DenseNet169 model has been fine-tuned and added sequentially to achieve better validation accuracy. From the validation, it is observed that it achieves an accuracy of 88.60% and outperforms other reported methods.
1. INTRODUCTION
Dementia is a general term utilized for memory-related neurological disorders; nonetheless, degenerative disorder is the most well-known significant neurocognitive impairment. As per the World Alzheimer's Report 2015, roughly 50 million individuals are experiencing dementia, where Alzheimer’s disease represents 70–80% of cases. Around millions of people worldwide live with dementia, indicating that there will be an exponential increase to more than 131 million by 2050. Alzheimer's disease is a neurodegenerative ailment that typically occurs in more established individuals and effectively annihilates human behaviors and retention. Neurodegenerative disease is currently the 6th leading source of death in the US. One in three patients dies from Alzheimer's disease or another dementia. An extensive study on magneto encephalogram (MEG) background activity from Alzheimer’s disease patients and elderly control subjects was carried out to identify the brain disorder [1]. There are numerous reasons for dementia. By and large, it results from the degeneration of neurons or aggravations in other body systems that influence the working of neurons [2, 3].
The neuroimaging data obtained from magnetic resonance imaging (MRI), diffusion tensor imaging (DTI), positron emission tomography (PET), and functional MRI (fMRI) are mostly used for analysis by physicians and radiologists. Alzheimer’s disease can be differentiated as very mild dementia, mild dementia, moderate dementia, and non-dementia. A few differences can be observed in the human brain structure during the four phases. The crucial phase for clinical trials is the mild and prodromal stages of Alzheimer’s disease. In order to delay Alzheimer’s disease deterioration, detection during the prodromal stage is the best way. It is not the illness of age, it is the sickness of the brain, and patients may show manifestations like trouble in tracking down the correct words, loss of memory, understanding what individuals are saying, and their ability to cope day to day tasks degrades concerning time. Presently, there is no remedy for most types of dementia; however, medicines, counsel, and support are accessible.
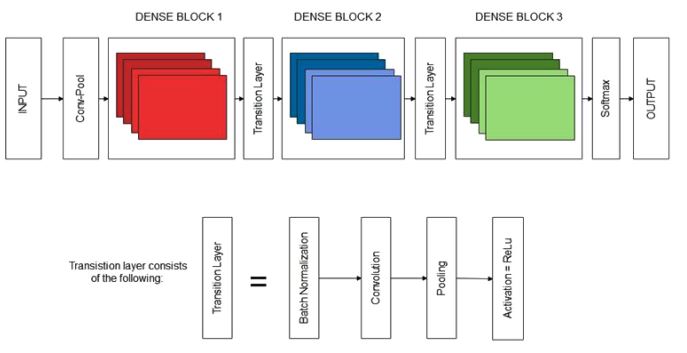
Due to the advancement in intelligence technology, many researchers have utilized the Machine Learning based approach for disease identification and classification [4, 5]. CNN had notable results over the previous decade in various fields identified with design acknowledgment, from voice recognition to image processing [6, 7]. The CNN-based auto-encoder is also used in various fields, such as, denoising, image classification, and pattern recognition [8, 9]. The new CNN architectures were created and tested using other CNN architectures: AlexNetet, VGG, DenseNet, and ResNet for Image classification [10]. The DenseNet has various advantages, such as it reduces the number of parameters and vanishing-gradient, inculcates feature reuse, and bolsters feature propagation. One of the significant advantages of DenseNets, besides parameter efficiency, is that it provides an improved course of gradients plus information during the network, which can be trained efficiently [11]. The DenseNet is one of the recent CNN architectures utilized in visual object recognition. The ResNet is considerably similar to the DenseNet; however, it holds key contrasts. The ImageNet and DenseNets datasets need lesser parameters than ResNet. Instead of using the summation, the DenseNet proposes concatenating outputs from the previous layers [12]. The name proposes that layers are entirely associated with the neurons in a network layer. Each neuron in a layer receives input from every neuron present in the past layer and hence, is densely connected. The dense layer is wholly associated, meaning every neuron in a layer correlates with those in the following layer [13].
Therefore, many researchers contributed to detecting and classifying Alzheimer’s disease in its early stage. The ensemble technique was employed to identify Alzheimer's disease genes using the sequence information of proteins, and classification was done based on the feature vectors using random forest [14]. Alickovic et al. [15] deployed the histogram to compute the feature vector from the MRI images, and finally, it was identified using the random forest algorithm. Furthermore, several multimodal deep-learning models to identify Alzheimer’s disease were proposed [16]. The denoising auto-encoder was used to extract the clinical features, and the 2D CNN was implemented to detect and classify Alzheimer’s disease. Oh et al. [17] proposed the volumetric CNN and utilized a learning-based method to classify and visualize Alzheimer’s disease from MR imaging. From the literature review, it is observed that regress research is still required to identify and classify Alzheimer’s disease in its early stage with more accuracy. The major contributions of the present research are as follows:
- Proposed the Sequential Convolutional Neural Network (S-CNN) to detect and classify Alzheimer’s disease in its early stage.
- Developed the model by combining the features of convolution operation and recursion operation to increase the detection and classification performance.
- Trained, validated, and tested the proposed S-CNN model using the ANSI and OASIS datasets.
- Validated the performance of the proposed S-CNN model using accuracy, F1 score, and precision and recall value.
2. MATERIALS AND METHODS
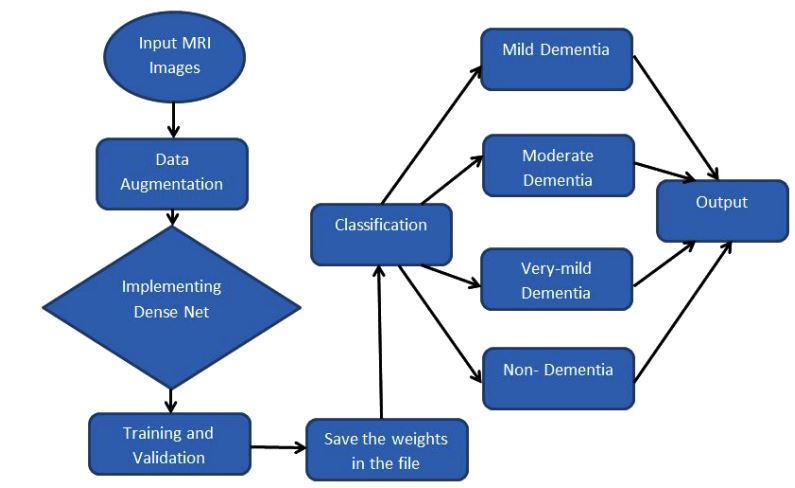
The overall workflow of the proposed sequential CNN model to detect and classify the various stages of Alzheimer’s disease is shown in Fig. (1). A sequential model is the simplest type of model, which uses the features of the convolution operation and recursion operation models.
2.1. Dataset
As mentioned earlier, the dataset examined in this paper contains normal and demented subjects. We split our dataset in order to formulate our model, which are as follows: Train, Validation, and Test. A total of 4098 images were used to train, 1023 images to validate, and 1367 images to test [18].
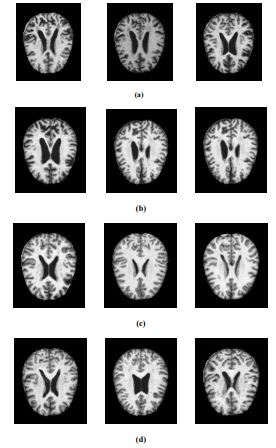
For this experiment, the Alzheimer's dataset [1] obtained from Kaggle is used. It is the combination of Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset and the Open Access Series of Imaging Studies (OASIS) dataset. The data has four classes of images, both in training as well as testing sets, as mild dementia, moderate dementia, non-dementia, and very mild dementia. This dataset consists of a cross-sectional collection of 6488 different subjects aged between 18 and 96 with 3 or 4 individuals per subject. Also, this dataset contains 46.7% and 53.3% as women and men subjects, respectively. The training and validation dataset consists of about 5,121 MRI images of sizes 224, 224, and 3. The pre-processed images are augmented by rotating, zooming, horizontal flip, and vertical flip [15]. The data collected includes mild dementia-717, moderate dementia-52, non-dementia-2560, and very-mild dementia-1792. The test dataset consists of 1,367 MRI images, including mild dementia-167, moderate dementia-12, non-dementia-640, and very-mild dementia-548. The MRI images of the various Alzheimer’s stage affected are extracted from the dataset and are illustrated in Fig. (2).
2.2. Pre-processing
One of the cardinal parts of deep learning is data pre-processing [19]. Before applying any algorithm for clinical studies, the pixel representation can be converted as useful for any imaging obtained. Hence, the pre-processing step plays a vital role in improving the performance of the proposed algorithm. Since we utilized the available dataset, image registration was used to align the data from the misaligned images. Since the MR scanning was very slow, MR slices within the same image sequence might likely be offset from one another. Therefore, the slice-level registration process was employed to align the MR images [11]. After the pre-processing, the data augmentation was done to produce an increased number of images with varying features, which helped train the model better. For data augmentation, we used a function called Image Data Generator, which produces tensor image data in batches, which is then augmented in real time. It creates a number of new images from an existing dataset with modifications in image parameters, such as rotation range, zoom range, rescales, horizontal flip, and vertical flip. The various parameters utilized for data augmentation are shown in Table 1.
2.3. Proposed Sequential CNN Model
The proposed sequential CNN model to detect and classify the various stages of Alzheimer's disease is shown in Fig. (3). We created the objects for training and validation that consisted of the respective images, as mentioned before. After that, we began with the initialization of our dense network using DenseNet169. In DenseNet, the interface connects every layer to another layer in a feed-forward fashion [20]. It is an architecture that exists in Keras, TensorFlow. The input shape for the network is 224,224,3, where 224 and 224 are the width and the height, respectively, and 3 is the number of input channels. The weights used were ImageNet. Moreover, we pre-trained our model on ImageNet by loading its file.
A model of the sequential type is created. It is a simple path to building a model in Keras. This allows us to construct our model layer after layer. The add() function was to build the layers. Then, our base model was incorporated, which is a dense net, into a newly created model. The weights of a pre-trained Densenet model were selected. All weights of all the layers were set to non-trainable from trainable, which is called freezing. During freezing, the state of the frozen layer will not be updated. The summary of the various layers incorporated in the proposed model is shown in Table 2.
| Rotation Range | 30 |
|---|---|
| Zoom range | 0.2 |
| Horizontal flip | True |
| Vertical flip | True |
| Rescale | 1 |
| Layer (Type) | Output Shape | Parameters |
|---|---|---|
| Densenet169 (Functional) | (None, 7, 7, 1664) | 12642880 |
| Dropout (Dropout) | (None, 7, 7, 1664) | 0 |
| Flatten (Flatten) | (None, 81536) | 0 |
| Batch_normalization | (None, 81536) | 326144 |
| Dense (Dense) | (None, 2048) | 166987776 |
| Batch_normalization_1 | (None, 2048) | 8192 |
| Activation (Activation) | (None, 2048) | 0 |
| Dropout_1 (Dropout) | (None, 2048) | 0 |
| Dense_1 (Dense) | (None, 1024) | 2098176 |
| Batch_normalization_2 | (None, 1024) | 4096 |
| Activation_1 | (None, 1024) | 0 |
| Dropout_2 | (None, 1024) | 0 |
| Dense_2 | (None, 4) | 4100 |
Trainable parameters: 169,259,268
Non-trainable parameters: 12,812,096
Next, we added the required elements to construct our model. The dropout layer was added to our model. During training, it randomly chose neurons and zeroed out their activation values [21]. It helps in preventing overfitting, which is most commonly an issue while constructing a model. Here, dropout is set at the rate of 0.5. It is one of the regularization methods used to prevent overfitting. In this study, each batch in its inputs was flattened using a flatten layer. In order to avoid covariate shift problems, layers of every input were added with a batch normalization layer. Dense was also added, which executed the activation functions. It connected the neurons from the previous layer and implemented operation activation. This was used several times in our model whenever required. The kernel initializer used was uniform, which is a constant variance scaling initializer. ReLu used in our study is an activation function. This helps in squashing the output to represent it comprehensively. At last, the model was added densely with an output space of 4 and a softmax activation function. It generated categorical probabilities, which could then be used to classify into four types.
3. RESULTS
3.1. Simulation Environment
The compilation of the model was done using the Adam optimizer at the learning rate of 0.001. In this study, the adequacy of Adam optimization is represented for Deep CNNs [22]. We computed the cross-entropy loss between the labels and predictions. One of the key issues in CNN is overfitting caused due to cross entropy's scale sensitivity and constant gradient updating [23]. A file was created to store the weights of the network during training. Our model used early stopping, which monitored the value of validation accuracy, and the training automatically stopped when its value stopped increasing. The various parameters used to simulate the proposed Sequential CNN model are tabulated in Table 3.
The patience set was 15 epochs, after which it stopped training if there was no improvement. The model only saved the best accuracy/latest values produced in an epoch to the weights file. Afterward, in the training process, we used the model to reduce the loss and improve the accuracy; over-training data regulated by the total number of epochs were used in the model to train [24].
| Parameter | Type/Range |
|---|---|
| Optimizer | Adam Optimizer |
| Learning Rate | 0.001 |
| CLR Policy | Exp_range with gamma = 0.99998 |
| Dropout | 0.5 |
| Batch Size | 128 |
| Number of epochs | 50 |
| Loss Function | Categorical Cross Entropy |
3.2. Experimental Results
The proposed S-CNN model was trained, validated, and tested for 20, 30, 40, and 50 epochs, and the obtained training accuracy, raining loss, validation accuracy, validation loss, testing accuracy, and testing loss are tabulated in Table 4. The number of epochs varied to check the performance each time, and the results produced by them were then compared.
| Epochs | Training accuracy | Training loss | Validation accuracy | Validation loss | Testing accuracy | Testing loss |
|---|---|---|---|---|---|---|
| 20 | 93.58% | 0.5945 | 81.95% | 1.2474 | 87.65% | 0.9885 |
| 30 | 95.68% | 0.4834 | 86.57% | 1.0276 | 87.48% | 1.1140 |
| 40 | 96.23% | 0.4533 | 84.59% | 1.2488 | 87.67% | 1.1491 |
| 50 | 96.81% | 0.4138 | 86.24% | 1.1191 | 88.60% | 1.1050 |
From the above table, it can be inferred that with an increasing number of epochs, the model gets trained better, thus increasing the training accuracy as well as the testing accuracy. It is observed that the accuracy of the proposed model is high in the training phase and becomes comparatively low in the testing phase. This may be due to the overfitting of the available data for the proposed model. In the future, it may be overcome by reducing the number of elements in the hidden layer, applying regularization, and adopting the dropout layers. The various performance metrics, such as recall, precision, and f1 score obtained during the training and testing phases, are shown in Table 5.
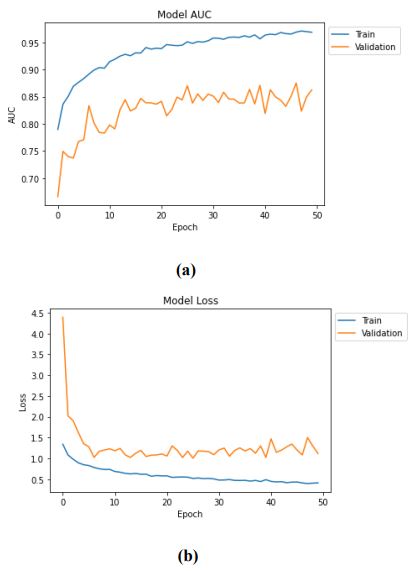
The metrics values mentioned above increase over a while, and the final value is presented. The results of the evaluation of loss and accuracy of the test dataset are mentioned in Table 4, where the values of f1, recall, and precision of the test dataset are obtained as 0.6518, 0.6487, and 0.6550, respectively. Therefore, it can be concluded that with 50 epochs, our model has given its best accuracy of 88.60%. The training and validation accuracy and loss of the proposed sequential CNN model are shown in Fig. (4).
| Evaluation Metrics | Training Phase | Testing Phase |
|---|---|---|
| Precision | 0.8534 | 0.6550 |
| Recall | 0.8256 | 0.6487 |
| F1 score | 0.8392 | 0.6518 |
4. DISCUSSION
The various samples applied to detect and classify the various stages of Alzheimer’s disease using the proposed Sequential CNN are shown in Fig. (5).
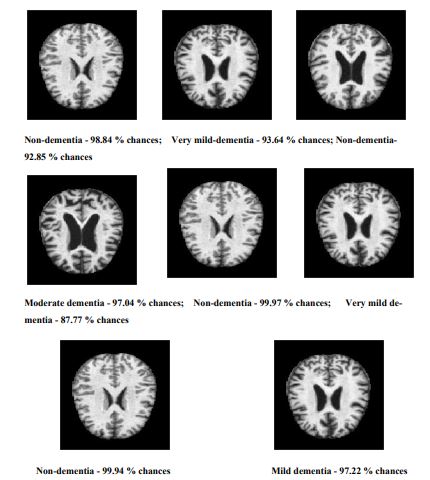
All the various stages, like dementia, non-dementia, moderate dementia, and very mild dementia, can classify with a minimum of 88.6% validation. The performance of the proposed method is compared with the existing methods, and it is tabulated in Table 6.
| Authors/Refs | Method/Model | Dataset | Validation Accuracy |
|---|---|---|---|
| Ullah, et al. [2] | CNN | OASIS | 80.25% |
| Neelaveni and Devasana [24] | SVM | - | 85% |
| Xu et al. [14] | Adaptive n-gram-k-skip model | UniProt database | 85.5% |
| Alickovic and Subasi [15] | Random forest | ADNI | 85.77% |
| Mohammed Abdullah et al. [25] | Decision tree | OASIS. | 73.33% |
| Venugopalan et al. [16] | 3D- CNN | ADNI | 86.04% |
| Oh et al. [17] | Volumetric CNN | ADNI | 78.77% |
| Proposed Method | Sequential CNN | ADNI & OASIS | 88.60 |
The various methods/models considered for the comparison are CNN [2], SVM [24], Adaptive n-gram-k-skip model [14], Random Forest [15], Decision Tree, 3-D CNN [16], and Volumetric CNN [17]. From Table 6, it can be observed that the proposed Sequential CNN model outperforms all other methods with a validation accuracy of 88.60%. This is due to the feature extracted and sequential nature of the CNN model.
CONCLUSION
In this study, the prognosis and prediction images of the degenerative disorder, also known as Alzheimer's disease, were compared using deep learning techniques. Over the decade, several diseases have been classified and identified using ML and DL methods. Based on affected regions and the aspects of parameters that determine the surface of the infected area, the images were classified and identified using neural approaches. In our proposed deep learning technique, DenseNet169-based S-CNN was used and compared with previous methods. It was observed that the proposed DenseNet169-based Sequential CNN architecture showed better classification accuracy than existing methods. The images were classified to get an ampler perception of the various stages of the disease. Alzheimer's disease is a brain-related issue that needs more categorization of testing and training sets of images from the dataset for the model to get trained and learned deeply. The research utilizing deep learning techniques is effective, and the development of 2D toward 3D CNN is essential, especially in Alzheimer’s disease.
LIST OF ABBREVIATIONS
| AD | = Alzheimer's Disease |
| MRI | = Magnetic Resonance Imaging |
| S-CNN | = Sequential Convolutional Neural Network |
| OASIS | = Open Access Series of Imaging Studies |
| MEG | = Magneto Encephalogram |
| DTI | = Diffusion Tensor Imaging |
| PET | = Positron Emission Tomography |
| fMRI | = Functional MRI |
| ADNI | = Alzheimer's Disease Neuroimaging Initiative |
| OASIS | = Dataset and Open Access Series of Imaging Studies |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The dataset available in Kaggle was used, which was approved for testing.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used in the studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data will be available upon reasonable request from the corresponding author [K.R].
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.


