All published articles of this journal are available on ScienceDirect.
SDF-1 Molecularly Imprinted Biomimetic Scaffold as a Potential Strategy to Repair the Infarcted Myocardium
Abstract
Background:
In situ cardiac tissue engineering aims to heal the infarcted myocardium by guiding tissue regeneration within the patient body. A key step in this approach is the design of a bioactive scaffold, able to stimulate tissue repair at the site of damage.
In the development of bioactive scaffolds, molecular imprinting nanotechnology has been recently proposed as a new functionalization strategy.
Objectives:
In this work, Molecularly Imprinted Particles (MIP) with recognition properties towards the stromal-derived factor-1 (SDF-1) were synthesized, characterized and used for the functionalization of a biomimetic scaffold. MIP are expected to favor the enrichment of the SDF-1 bioactive molecule within the scaffold, thereby promoting myocardial regeneration.
Methods:
MIP were obtained by precipitation polymerization, using the SDF-1 molecule as a template. Alginate/gelatin/elastin sponges were fabricated by freeze-drying and functionalized by MIP deposition. Morphological, physicochemical and functional analyses were performed both on MIP and on MIP-modified scaffolds. A preliminary biological in vitro investigation was also carried out using rat cardiac progenitor cells (rCPCs).
Results:
Imprinted nanoparticles with an average diameter between 0.6 and 0.9 µm were obtained. Infrared analysis of MIP confirmed the expected chemical structure. Recognition and selectivity tests showed that MIP were able to selectively recognize and rebind the template, even after their deposition on the scaffold. In vitro biological tests showed that cell adhesion to the scaffold was promoted by MIP functionalization.
Conclusion:
Results obtained in the present study suggest that biomimetic alginate/gelatin/elastin sponges, functionalized by MIP with recognition properties towards SDF-1, could be successfully used for tissue engineering approaches to repair the infarcted heart.
1. INTRODUCTION
Myocardial Infarction (MI) is the segmental loss of cardiac tissue resulting from the obstruction of major coronary arteries. The human heart possesses a limited capacity to regenerate itself, because most adult cardiac myocytes are terminally differentiated and have a reduced ability to replicate after injury [1]. Therefore, heart failure following a heart attack is caused by a substantial death of cardiomyocytes in the affected area that cannot be replaced by new functionally competent cells. The best therapeutic option is heart transplantation, which is limited by the low number of donors and immune-mediated adverse events, leading to organ rejection after transplantation. As a result, in recent years, considerable interest is grown in developing alternative therapeutic strategies and, in particular, in cardiac tissue engineering.
Cardiac tissue engineering aims at the repair, regeneration and replacement of damaged cardiac tissue, by implanting a bioactive biomaterial directly at the injured site, in a seeded or unseeded configuration. In particular, the in situ regeneration of cardiac muscle includes the mobilization of progenitor or stem cells to the damaged area and the subsequent stimulation of a regenerative program aimed at restoring tissue integrity [1, 2]. Recent studies have suggested that the heart contains resident stem cells that can be induced to develop new cardiac and vascular tissue. To exploit this knowledge, the in situ approach for cardiac tissue regeneration is based on the implantation of a preformed or injectable bioactive scaffold, properly interacting with the recruited endogenous cells [1-3]. Consequently, the in situ approach includes two key steps: the identification of suitable bioactive molecules and the consequent choice of an appropriate scaffold functionalization method.
Molecules used for scaffolds functionalization include growth factors, extracellular matrix proteins and peptides, antibodies, aptamers and angiogenic factors [2]. Each group of bioactive molecules requires a different functionalization strategy that can be traditionally chosen among direct absorption, covalent attachment, surface deposition and bulk functionalization [2].
In particular, myocardial regeneration can be stimulated by the delivery of cytokines that induce the recruitment of stem cells which, upon engraftment and differentiation toward myocardial phenotypes, lead to tissue growth. Among these cytokines, SDF-1 is the most potent chemoattracting agent for stem cells that possess the specific receptor CXCR4, including cardiac progenitor cells (CPCs) [4]. SDF-1 also stimulates cell retention, growth and proliferation [5-7]. In addition, it has been reported to decrease apoptosis and increase cell survival in the infarct zone [6, 7], as well as to improve heart function and survival after MI [8-10].
Traditionally, growth factors are physically loaded into the scaffolds, directly in the scaffold matrix or through the use of carriers. They can be released from the scaffold and trigger or modulate new tissue formation. For example, the sustained release of SDF-1 from a modified hyaluronic acid hydrogel matrix, during hydrolytic degradation of the scaffold, was proven effective for the treatment of MI, thanks to the efficient recruitment of bone marrow stem cells [11]. Another research group developed biomimetic injectable hydrogels containing SDF-1 and angiogenic peptides to promote respectively stem cell homing and angiogenesis and they reported improved left ventricle function, increased angiogenesis, wall thickening and decreased infarct size within the infarcted region [12]. In another study, SDF-1 was loaded into nanofibers formed from self-assembling peptides. The sustained release of SDF-1 enhanced stem cells recruitment and improved cardiac function post-MI in rats [13]. An innovative approach to produce advanced bioactive scaffolds for tissue engineering is represented by molecular imprinting technology [14]. This technology permits the introduction, into a polymeric material, of recognition sites for a specific molecular species (called template) through the polymerization of a monomer in the presence of the template [15]. The spatial arrangement of the cavities, created around the template during polymerization, is maintained by the polymer after template extraction and confers a selective “memory” towards the template molecule. The most extensively studied applications of molecularly imprinted polymers include separation by affinity sector, synthetic antibodies, biomimetic sensors and intelligent polymers [16-18]. Attractive developments in the biomaterial field in general and in tissue engineering, in particular, can also be envisaged.
Our research group proposed for the first time the realization of polymeric materials for applications in tissue engineering, which operate as “smart” systems, thanks to the presence on their surface of molecularly imprinted polymers with recognition properties towards molecules of interest that are able to guide and control cellular processes such as recruitment, adhesion, proliferation and differentiation. The feasibility of this approach has already been investigated for the production of bioactive scaffolds with recognition properties towards peptide sequences from the extracellular matrix proteins (such as fibronectin [19, 20] and laminin [20, 21]).
In this work, we report on the synthesis and characterization of Molecularly Imprinted Particles (MIP) with recognition properties towards SDF-1 (hereafter indicated as MIP-SDF1) to be used for scaffold functionalization. After verifying the recognition and rebinding capability towards SDF-1, synthesized particles were used to modify biomimetic sponges based on a blend of alginate, gelatin and elastin (AGE). These scaffolds were prepared and investigated by us previously [20], with the aim to mimic the composition and the interactions of the native cardiac extracellular matrix. It is in fact well known that the extracellular matrix not only provides mechanical support, but also transduces essential molecular signals in healthy and diseased myocardium. Specifically, its composition plays an important role in cardiac regeneration and repair after cardiac injury [22, 23].
Therefore, the combination of biomimetic alginate/gelatin/elastin sponges with molecular imprinted particles could have a synergistic effect towards cardiac repair: in addition to provide a native-like environment, functionalization by deposition of MIP-SDF1 is expected to allow an enrichment on the scaffold of SDF-1, to promote the migration and retention of stem cells at the site of injury. Functionalized sponges underwent a morphological, physicochemical and functional characterization. In addition, a preliminary in vitro biological investigation was performed to verify their effect on rat CPCs (rCPCs).
2. MATERIALS AND METHODS
2.1. Materials
Methacrylic acid (MAA>99%) from Sigma Aldrich (Milan, Italy) was purified by vacuum distillation to remove the polymerisation inhibitor. Trimethylpropane trimethacrylate (TRIM) and azobis(isobutyronitrile) (AIBN>98%) from Sigma Aldrich were used as supplied. SDF-1 and Insulin-like growth factor 1 (IGF-1) were provided by Immunological and Biochemical TestSystems (Binzwangen, Germany). Acetonitrile (ACN>99.9%) from Carlo Erba Reagenti (Milan, Italy) was of HPLC grade purity. Phosphate buffered saline (PBS) was from Sigma Aldrich. Alginate (viscosity of 2% solution at 25°C = 250 cps), gelatin (type B from bovine skin), elastin (from bovine neck ligament) and glutaraldehyde (GTA, 25% aqueous solution) were acquired from Sigma Aldrich. Calcium chloride was purchased from Carlo Erba Reagenti (Milan, Italy).
2.2. MIP-SDF1: Synthesis Procedure
MIP-SDF1 were prepared using the precipitation polymerisation method described in previous papers [19-21], at the composition shown in Table 1.
Briefly, the template molecule was dissolved in a 95/5 (v/v) ACN/bi-distilled water (ddH2O) solution and introduced into borosilicate glass tubes. The functional monomer, the cross-linker and the initiator were added to the polymerisation solution. The tubes were sealed under dry nitrogen. The polymerization was thermally initiated at 60°C and carried out for 20 h, under constant stirring. At the end of the polymerization process, the tubes were left to settle under the hood overnight. The residual polymerization solution was then separated from the particles and subjected to chromatographic and spectrophotometric analysis. Collected particles were washed with ACN/ddH2O mixture to remove residual monomer, initiator, cross-linker and not entrapped template, and dried in the oven at 37°C. Washing waters were collected for subsequent chromatographic and spectrophotometric analysis. Dried particles were then crushed in a mortar to avoid the formation of aggregates.
The last step of MIP preparation was the extraction procedure to remove the template molecule from imprinted particles and create the recognition sites. Template extraction was performed by repeated washing with PBS, under vigorous stirring. Three washing cycles were performed. At the end of each washing, the sample was centrifuged and the collected supernatant underwent spectrophotometric analysis, using the QuantiPro BCA assay kit (Sigma Aldrich, Milan, Italy), to quantify the amount of extracted template.
Control particles (CP) were prepared with the same procedure as the imprinted ones but in the absence of the template molecule.
| Template (µg) | MAA (µl) | TRIM (µl) | AIBN (µg) | ACN/ddH2O 95/5 (ml) | |
|---|---|---|---|---|---|
| MIP-SDF1 | 20 | 170 | 640 | 12 | 30 |
| CP | - | 170 | 640 | 12 | 30 |
2.3. Preparation of MIP-SDF1 functionalized AGE sponges
AGE sponges were prepared following a procedure established by us in previous work [20]. Briefly, alginate, gelatin and elastin 2% (w/v) bi-distilled water solutions were prepared separately at 50°C.
Different volumes of natural polymer solutions were mixed together, under stirring at room temperature, in order to obtain an AGE blend with a A:G:E=10:80:10 weight ratio.
A known volume of the blend was poured into polystyrene Petri dishes and freeze-dried. The obtained sponges were then treated with GTA vapours at 37°C for 18 hours for proteins cross-linking, and then immersed for 30 minutes in a solution containing 2% (w/v) CaCl2 in bi-distilled water for polysaccharide cross-linking.
After cross-linking treatments, samples were immersed for 16 hours in a coagulation bath (0.5 M acetic acid) to promote ionic interactions among the components.
Since GTA and acetic acid could be dangerous for cells, cross-linked sponges were washed repeatedly with bi-distilled water to remove excess GTA and acetic acid, until UV-spectrophotometric and pH analysis of wash waters did not reveal any residual trace.
At the end of the washing procedure, the scaffolds were freeze-dried again.
Sponges were functionalized by deposition of MIP on their surface. A known amount of MIP was dispersed in PBS and deposited on preformed scaffolds. After MIP deposition, sponges were freeze-dried. Sponges modified with CP were used as controls.
2.4. Morphological Analysis
The morphology of imprinted and control particles and AGE sponges, before and after functionalization, was examined by a JSM 5600 scanning electron microscope (SEM, Jeol Ltd, Tokyo, Japan). Before analysis, dried samples were mounted on metal stubs and coated with a 24 Kt gold layer.
2.5. Chromatographic and Spectrophotometric Analysis
The amount of residual monomer in the polymerization solution and in the washing waters was quantified by high-performance liquid chromatography (HPLC, 200 Series HPLC system, Perkin Elmer, Waltham, Massachusetts, United States), with a UV/VIS detector to determine the percentage of monomer conversion during MIP synthesis. An Alltima C18 column (250 mm length × 4.5 mm i.d.) was used. The mobile phase was a solution ACN/ddH2O (80/20 v/v). The flow rate was set at 1 ml/min for 15 mins. The injection volume was 100 µl. The detector wavelength was set at λ= 210 nm.
The percentage of template entrapped by the polymeric particles and the percentage of extracted template were determined by spectrophotometric analysis. Template entrapment was evaluated by measuring the residual template amount in the polymerization solution and washing waters. Template extraction was investigated by quantifying the template amount in the extraction solution. A QuantiPro BCA assay kit was used for template quantification. In particular, the amount of SDF-1 was estimated, measuring spectrophotometrically the optical density of the samples at λ=562 nm, against a calibration curve, using bovine serum albumin as per the manufacturer’s protocol.
2.6. Infrared Spectroscopy
Fourier Transformed Infrared Spectroscopy (FT-IR) spectra were recorded on a Spectrum-One instrument (Perkin Elmer, Waltham, Massachusetts, United States). The spectrometer was equipped with Attenuated Total Reflectance (ATR) objective lens with a penetration depth of less than 1 µm. All spectra were obtained at 4 cm-1 and represented the average of 16 scans. Spectral images were also acquired using the infrared imaging system of the instrument (Spotlight 300, PerkinElmer, Waltham, Massachusetts, United States). The spectral resolution was 4 cm-1. The spatial resolution was 100 × 100 µm.
2.7. Recognition and Selectivity Tests
Recognition and selectivity tests were carried out for MIP-SDF1 and relative control, (CP).
To evaluate the rebinding capacity, three samples of both MIP and CP (10 mg each) were put in contact with 1 ml of a rebinding solution, consisting of SDF-1 in PBS, at a concentration of 0.125 µg/ml. The tubes containing the rebinding solution and MIP or CP, were maintained under constant stirring for 30 minutes. The supernatant was separated from the polymer by centrifugation for 30 min at 14,000 rpm. The procedure was repeated three times, replacing every time the supernatant with fresh rebinding solution. The residual SDF-1 concentration in the rebinding solution was determined by spectrophotometric analysis using the QuantiPro BCA assay kit.
The amount of template rebound by MIP was calculated as follow:
Quantitative binding (µmol of template)=V×(Ci-Cf)
where V, Ci and Cf represent, respectively: the volume of the rebinding solution, the initial rebinding solution concentration and the rebinding solution concentration after recognition.
The imprinting effect was evaluated through the recognition factor (α), calculated according to the following equation:
αMIP/CP = template rebound by MIP/template rebound by CP
When the value of the recognition factor is higher than one, the recognition capability of MIP is demonstrated.
Selectivity was also verified by incubating fixed amounts of both MIP and CP with 0.125 µg of a template analogue (IGF-1) in 1 ml of PBS and then proceeding as for rebinding experiments.
The selectivity factor (α’) and the specific selectivity factor (α”) were calculated as indicated in the equations below:
α'template/analogue = template bound by MIP/analogue bound by MIP
α” template/analogue=(template bound by MIP-template bound by CP)/(analogue bound by MIP-analogue bound by CP)
Particles selectivity is demonstrated when α’ and α” are higher than 1.
Recognition experiments were also performed on MIP-modified sponges to evaluate if the rebinding capacity was maintained by MIP after deposition on the scaffolds. The procedure followed was similar to that used to test MIP recognition: three samples (1 cm2 each) of MIP-modified sponge were put in contact with 1 ml of the rebinding solution. At the end of each rebinding cycle, the rebinding solution was collected and centrifuged before spectrophotometric analysis. Three samples (1 cm2 each) of scaffolds modified with CP were used as controls.
2.8. In vitro Biological Characterization
2.8.1. Rat Cardiac Progenitor Cells Isolation and Culture
rCPCs were isolated from Wistar rats and cultured as previously described [24-26]. For the experiments described in this study, rCPCs at passages 4 and 5 (P4–P5) were employed.
2.8.2. DiI Cell Labeling
In order to better detect rCPCs cultured on the sponges, before seeding, cells were stained with CellTracker CM-DiI (Invitrogen, C-7001) according to manufacturer instructions [27].
2.8.3. Cell Culture on AGE Scaffolds
AGE sponges (functionalized with MIP-SDF-1 or containing CP, as control) were sterilized by UV exposure for 15 min on each side and cells were seeded on their surface at 45 × 103 cells/cm2 density, according to a previously described methodology [28].
CM-DiI labelled rCPCs seeded on AGE scaffolds with CP or MIP-SDF1 were evaluated at 2 and 14 days after plating. Quantification of cell adhesion was performed by computing the fractional area occupied by CM-DiI fluorescent rCPCs using “Image Pro Plus 4.0” software [27].
3. RESULTS AND DISCUSSION
3.1. MIP Synthesis
MIP synthetized in this work are made by a polymeric matrix of poly(methacrylic acid) (PMAA) and were obtained by using a template molecule of protein type (SDF-1). The synthesis was performed by radical polymerization, in a solution of ACN/ddH2O 95/5 (v/v), in the presence of TRIM, as crosslinker, and AIBN, as initiator.
The presence of water in the polymerization solution has a strong influence on the morphology of the rebinding sites and consequently on the rebinding performance of MIP, because of its polar nature, which influences non-covalent interactions between the monomer and the template molecule during polymerization [29]. Therefore, the amount of water used in the polymerization solution was determined as the minimum one necessary to dissolve the template.
The reaction yield was 94.7% for MIP and 82.2% for CP. This result could be explained by the establishment of interactions between the monomer and the template, which could favour a higher polymerization yield.
3.2. MIP Characterization
3.2.1. Morphological analysis
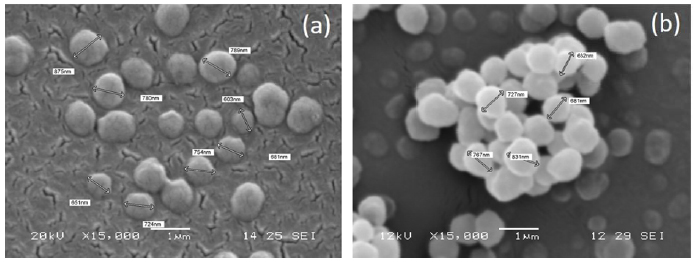
SEM micrographs were acquired for MIP-SDF1 and relative CP (Fig. 1). MIP particles were well separated, with a spherical shape and homogeneous, with a diameter ranging between 600 nm and 900 nm. No morphological difference was observed between MIP and CP.
3.2.2. Chromatographic and Spectrophotometric Analysis
The percentage of monomer conversion for MIP and CP was evaluated by HPLC analysis, determining the amount of residual monomer in the polymerization solution, collected at the end of the polymerization process and in the washing waters. Results are shown in Table 2.
Monomer conversion was lower for CP than for MIP, in agreement with the lower reaction yield.
The amount of template contained in the final product was calculated from the difference between the amount of template contained in the polymerization solution before polymerization and the residual amount of template present both in the polymerization solution after polymerization and in the washing waters. The spectrophotometric analysis of both polymerization solution and washing waters, performed using the QuantiPro BCA assay kit, did not reveal any trace of residual SDF-1. This result demonstrated that all the template introduced in the reaction solution was entrapped by MIP.
The template molecule was then removed by repeated washings with PBS, under vigorous stirring. The extraction procedure was repeated for three times on each sample and the extraction solution collected at the end of each extraction cycle was analyzed by QuantiPro BCA assay kit to quantify the amount of extracted template. As reported in Table 2, the removal of template from the particles was high, even if not complete. This result could be explained by the high cross-linking degree of MIP and therefore, consequent rigidity of the matrix, which can determine increased template retention. Moreover, the possible establishment of hydrogen bond interactions between the functional monomer and the template molecule could obstacle template removal. It is also well known in the literature that the removal of a protein template from imprinted polymers is challenging due to protein high molecular weight, which retards diffusion through a dense polymer network [30]. Percentages of template removal varying between 50% and 95% have been reported in the literature for protein-imprinted polymers [31-35]. Therefore, the percentage of template removal obtained in this work is similar and sometimes higher than those reported in the literature for other protein templates.
| - | Monomer conversion (%) | Entrapped template (%) | Extracted template (%) |
|---|---|---|---|
| MIP-SDF1 | 93.4 | 100.0 | 73.0 |
| CP | 81.1 | - | - |
3.2.3. Infrared Spectroscopy
Chemical maps were acquired for MIP-SDF1 (before template extraction) and relative CP. From the chemical map, the medium spectrum (which is the most representative spectrum of the chemical map) was recorded (Fig. 2). Comparing the medium spectrum of MIP-SDF1 with that of CP, differences were observed in the region between 1700 and 1500 cm-1, attributable to the adsorption bands due to Amide I and Amide II, which are typical of the template, SDF-1, which has a protein nature. In particular, in the medium spectrum of MIP-SDF1 it was possible to observe an increase of intensity for the band at 1630 cm-1 (due to Ammide I) and an adsorption peak at 1558 cm-1 (due to Ammide II).
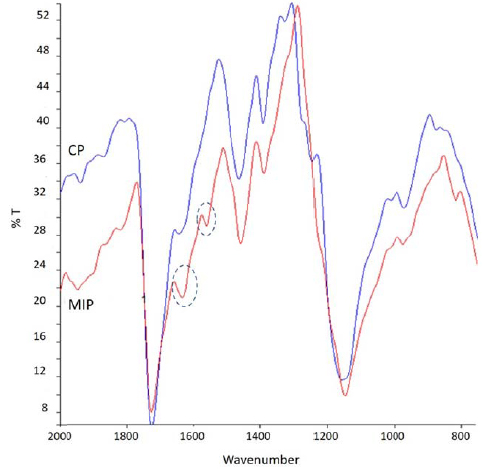
In both spectra, the adsorption band at 1725 cm-1 was well evident. This band was due to ester C=O of the PMAA polymeric matrix and it was considered diagnostic to evaluate later the presence of MIP on functionalized sponges.
Spectra of MIP-SDF1 and CP did not show differences in other regions of adsorption, evidencing the same chemical structure of MIP and CP, with the only exception of the presence of protein template in MIP.
3.2.4. Recognition and Selectivity Tests
The aim of the recognition tests is to determine the ability of imprinted particles to recognize and rebind the template molecule and also to verify if this rebinding ability is due only to a not specific rebinding (due to the polymeric matrix) or to a specific rebinding, due to nanocavities which are present on imprinted nanoparticles after the extraction process. Results of the recognition test, performed as described in paragraph 2.7, are shown in Table 3.
The recognition factor (α), calculated as the ratio between the total amount of template rebound by MIP (TMIP) and the total amount of template rebound by CP (TCP), was equal to 1.59. The recognition factor higher than 1 shows that MIP were able to recognize and rebind the template better than control.
We also evaluated the specific rebinding (RS), as the difference between TMIP and TCP, which was equal to 0.40 μg of template molecule. From RS, we calculated the percentage of rebinding, as the ratio between RS and TEXTR, where TEXTR is the amount of template (in µg) extracted from MIP which underwent the rebinding test, corresponding to 0.54 µg. The percentage of rebinding resulted therefore 74%.
Overall, these results showed that MIP have a rebinding ability higher than CP, due to the formation of specific recognition sites in their polymeric matrix.
Selectivity tests were also carried out with the aim to verify the selective rebinding of MIP towards SDF-1, with respect to rebinding of an analogue molecule. The molecule used as analogue was the growth factor IGF-1 and the procedure followed is that described under paragraph 2.7. Results are collected in Table 3.
The selectivity factor (α’), calculated as the ratio between the total amount of template molecule rebound by MIP and the total amount of analogue molecule rebound by MIP, was 2.40.
The specific selectivity factor (α”) was also calculated, as the ratio between (template bound by MIP-template bound by CP) and (analogue bound by MIP-analogue bound by CP). It resulted equal to 2.22.
Both the selectivity factor and the specific selectivity factor were therefore much higher than one, showing that MIP were also able to distinguish between the template and an analogue molecule.
| Template | Analogue | ||||
|---|---|---|---|---|---|
|
Quantitative binding (µg) |
Recognition factor (α) |
Quantitative binding (µg) |
Selectivity factor (α’) | Specific selectivity factor (α”) | |
| MIP-SDF1 | 1.08 | 1.59 | 0.45 | 2.40 | 2.22 |
| CP | 0.68 | 0.27 | |||
3.3. Characterization of MIP Functionalized AGE Sponges
3.3.1. Morphological Analysis
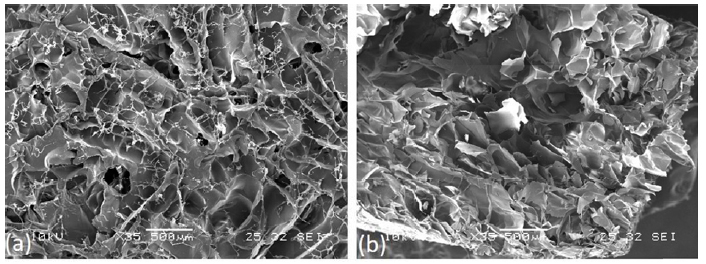
Morphological analysis of AGE sponges before and after deposition of MIP and CP was performed by SEM (Fig. 3). Micrographs were acquired to analyze surface and section of not functionalized AGE sponge, AGE sponge functionalized with MIP and AGE sponge with CP.
As already observed by us in a previous paper [20], AGE sponges showed a highly porous structure with well-interconnected pores. MIP and CP nanoparticles appeared homogeneously distributed on AGE sponges. No significant difference was observed between sponges functionalized with MIP or CP, neither between functionalized and not functionalized sponges.
3.3.2. Infrared Analysis
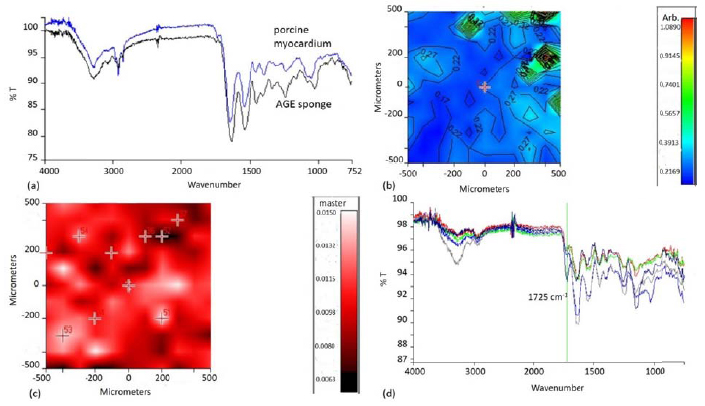
A chemical map of the AGE sponge was acquired and from it, the medium spectrum was recorded (Fig. 4a). The following adsorption peaks, typical of the three natural polymers used for the blend, can be observed:
- Between 4000 and 3000 cm-1, due to N-H and O-H groups;
- Between 2900 and 2700 cm-1, due to symmetric and asymmetric stretching of C-H and C-H2 groups;
- At 1635 cm-1, due to C=O group of Ammide I;
- At 1545 cm-1, due to C-N and N-H groups of Ammide II;
- At 1030 cm-1, due to polysaccharidic C-O-C.
These results were in agreement with what was already observed in our previous paper [20].
As the aim of using a three component blend in the preparation of the sponges was to mimic the chemical composition of native ECM, the medium spectrum of AGE sponges was compared with that of a healthy pig myocardium (Fig. 4a). The two spectra appeared very similar: they showed analogies both for the presence of typical adsorption peaks, in particular those due to protein and polysaccharidic components, and in terms of intensity and frequencies of adsorption.
To further characterize similarities between AGE sponges and native tissue, the band ratio between the typical polysaccharide adsorption peak and the typical protein adsorption peak was calculated (Fig. 4b). This value was in the range 0.22-0.27. The same analysis was carried out on a sample of healthy porcine myocardium and the band ratio was equal to 0.29. This result further confirmed that our attempt to produce a biomimetic scaffold mimicking the chemical composition of native myocardium was successfully achieved.
Chemical imaging investigation was repeated on the sponges after the deposition of MIP. As previously pointed out, MIP were characterized by a typical adsorption peak at 1725 cm-1. Therefore, the chemical map of MIP functionalized AGE sponges, in function of this adsorption band, was acquired (Fig. 4c). The map showed the presence of globular structures attributable to particles aggregates. All the spectra acquired in correspondence of these globular structures (Fig. 4d) showed a well-evident adsorption peak at 1725 cm-1, confirming the successful deposition of MIP on AGE sponges.
3.3.3. Recognition Experiments
The ability of synthetized MIP to recognize and rebind the template molecule (SDF-1), after deposition on AGE sponges, was evaluated by performing recognition tests on AGE sponges functionalized with MIP-SDF1. Sponges containing CP were used as control. Results are shown in Table 4.
| Template | ||
|---|---|---|
|
Quantitative binding (µg) |
Recognition factor (α) | |
| Sponge + MIP-SDF1 | 0.34 | 1.31 |
| Sponge + CP | 0.26 | - |
The recognition factor, calculated as the ratio between total template quantitative binding of the sponges functionalized with MIP and total template quantitative binding of the sponges containing CP, was equal to 1.31. This result showed that the functionalized scaffold was able to recognize and rebind the template molecule, thanks to the specific nanocavities created on MIP after template extraction.
Values of quantitative binding were lower than those obtained for free nanoparticles. In fact, for free nanoparticles we obtained a total quantitative binding of 1.08 µg SDF-1, performing three rebinding cycles on three samples of 10 mg each (Table 3). In the case of functionalized sponges, a total quantitative binding of 0.34 µg SDF-1 was obtained performing three rebinding cycles on three samples, each one containing 5 mg of MIP (Table 4). This could be explained by the fact that when imprinted nanoparticles are deposited on the sponges, not all the recognition nanocavities remain easily accessible for the rebinding process, considering also the large dimension of the protein template. However, the amount of rebound SDF-1 can be considered sufficient to promote a desired cellular response. In fact, as reported in the literature, an amount of SDF-1 between 50 and 500 ng has been demonstrated able to promote stem cell recruitment and retention [12, 36].
3.3.4. In Vitro Biological Characterization
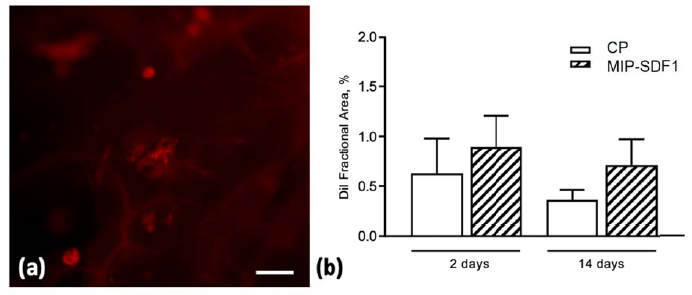
The quantitative evaluation of DiI labelled cells cultured on AGE sponges documented that MIP-SDF1 functionalization was able to increase by 20% and 45% rCPC adhesion to biomimetic scaffolds, at 2 and 14 days after cell seeding, respectively (Fig. 5).
CONCLUSION
The development of a biomimetic and bioactive scaffold is a key step to promote in situ myocardial tissue regeneration after an ischemic injury. For this purpose, in this work, we investigated the innovative combination of a biomimetic scaffold material, based on a blend of three natural polymers, with a new functionalization strategy, based on molecular imprinting. MIP with recognition properties towards SDF-1, a stem cell homing factor, were synthesized and subsequently deposited on preformed sponges, with the aim to obtain bioactive scaffolds able to favor a local enrichment of SDF-1 and consequently to promote CPCs migration and retention at the site of injury, overall enhancing myocardium healing. Results of the morphological, physicochemical and functional characterization confirmed the obtainment of a scaffold material mimicking the chemical composition of native cardiac ECM, as well as the synthesis of imprinted nanoparticles able to selectively recognize and rebind the SDF-1 template, even after deposition on the sponges. Results of a preliminary biological characterization using rCPCs were also encouraging, showing that cell adherence and retention on the scaffolds were promoted by MIP functionalization. In conclusion, the innovative approach based on the combination of a biomimetic scaffold with an innovative application of molecular imprinting as functionalization strategy could be a valid therapeutic option endowed with the potential to treat the infarcted heart by in situ tissue engineering. Future developments will include further biological characterization, through in vitro and in vivo tests, as well as the fabrication of scaffolds with structures more closely mimicking the architecture of native ECM.
LIST OF ABBREVIATIONS
| ACN | = Acetonitrile |
| AGE | = Alginate/Gelatin/Elastin |
| AIBN | = azobis(isobutyronitrile) |
| ATR | = Attenuated Total Reflectance |
| CP | = Control Particles |
| CPCs | = Cardiac Progenitor Cells |
| ddH2O | = bi-distilled water |
| FT-IR | = Fourier Transformed Infrared Spectroscopy |
| GTA | = Glutaraldehyde |
| HPLC | = High-Performance Liquid Chromatography |
| IGF-1 | = Insulin-like Growth Factor 1 |
| MAA | = Methacrylic Acid |
| MI | = Myocardial Infarction |
| MIP | = Molecularly Imprinted Particles |
| MIP-SDF1 | = Molecularly Imprinted Particles with recognition properties towards SDF-1 |
| PBS | = Phosphate Buffered Saline |
| PMAA | = poly(methacrylic acid) |
| rCPCs | = rat cardiac progenitor cells |
| SDF-1 | = stromal-derived factor-1 |
| TRIM | = Trimethylpropane trimethacrylate |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used in this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The authors confirm that the data supporting the findings of this research are available within the article.
FUNDING
This research was partially supported by European Commission FP7 Programme (Grant No. 214539).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.


