All published articles of this journal are available on ScienceDirect.
An Improved Segmentation and Classifier Approach Based on HMM for Brain Cancer Detection
Abstract
Introduction:
Brain tumors are fatal diseases that are spread worldwide and affect all types of age groups. Due to its direct impact on the central nervous system, if tumor cells prevail at certain locations in the brain, the overall functionality of the body is disturbed and chances of a person approaching death are high. Tumors can be cancerous or non-cancerous but in many cases, the chances of complete recovery are less and as a result death rate has increased all over the world despite recent advancements in technology, equipment and awareness. So the main concern is to detect brain related diseases at early stages so that they do not spread into vital parts of brain and disrupt body functions. Also, more precise and accurate technologies are required to serve as aid in the diagnosis, treatment and surgery of brain.
Aims & Objectives:
Therefore, its high alarming time to monitor mortality statistics and develop faster and accurate methods to curb the situation by simulating tissue deformation and locating cancerous nodes which is currently the prominent area of interest.
Methods:
A brain tumor is used to design the deformation model. Early stage detection of tumors is difficult from images. Moreover, the accuracy involved is low. Keeping all this into consideration, a machine learning approach has been developed for classification of cancerous and non-cancerous tissues so that the tissues having risk of future problem can also be recognized. The patient’s deformation model can be designed and brain tumor patterns are given as input on the basis of which tumor in the brain is marked. The proposed method of segmentation is based on a statistical model called Hidden Markov Model (HMM) which extricates the cancerous portion out of fed input MRI image along with the calculation of parameters such as Peak Signal-to-Noise Ratio (PSNR), Mean Square Error (MSE), fault rate dust detection and accuracy.
Results &Discussion:
The results obtained from parametric analysis show that HMM has performed better than the technique of Support Vector Regression (SVR) for brain cancer segmentation in terms of PSNR, MSE, fault rate dust detection and accuracy. So image processing is used in combination with Hidden Markov Model for classification and analysis to which MRI images are fed as input.
Conclusion:
In this way, integration of artificial intelligence techniques with image processing can serve as a good way for segmentation of tumors and for classification purposes with good accuracy.
1. INTRODUCTION
The brain is an extremely vital part of living beings that is being responsible for curbing and collaborating the cell activi ties in human body. Many of the major bodily functions are performed by the brain like control of motor movements of different parts of body, activation of body senses in organs (ears, nose, eyes, tongue, skin), maintenance of rhythmic cycles of pulse, breathing and cardiac rates [1] controlling the way language is understood and expressed by living beings, coordination and communication between different parts of body with consecutive actions, control of emotions and expressions [2], choice of actions and behaviour through intelligence to accomplish the set targets etc. So the nervous system incorporates the brain, spinal cord and nerve fibres that are responsible for supervising the overall functioning of the entire body and provide a full-fledged control over the body parts through communication carried out by transmission and reception of signals from one part to the other. The brain is the most complex human organ constituting a network of billions of nerves and their cells called neurons that simultaneously interact through transmission and reception of messages with one another to control and coordinate functions of human body. The messages are in the form of electrochemical processes that constitute electrical current due to the movement of ions (chloride ions etc.) in the body. Such biological neural networks [3] that are formed through complicated connections of billions of cells and nerves form the base of artificial neural networks. Tumors are crucial manifestation of an immense set of diseases called lumps or cancers. These tumors are difficult to diagnose in medical images particularly in the early stages [4]. Cerebrum tumors can be primary or secondary depending on the site of tumor creation. Primary tumors originate in the brain while secondary tumors originate in varied parts of the body and then reach to the brain by spreading. Brain cancer can be detected using image segmentation techniques [5], image enhancement techniques or morphological techniques [6].
2. OBJECTIVES
The fundamental idea behind the thesis work is to detect brain cancer in the input MRI and classify brain images into cancerous and non- cancerous cells. The objectives of this thesis are given below:
- To propose and implement an improved segmentation and classifier approach based on HMM for brain cancer detection.
- To compare the results obtained with previous technique.
3. METHODOLOGY
The objectives specified above are achieved through following procedure.
- Step-1: Study of literature based on brain cancer detection techniques.
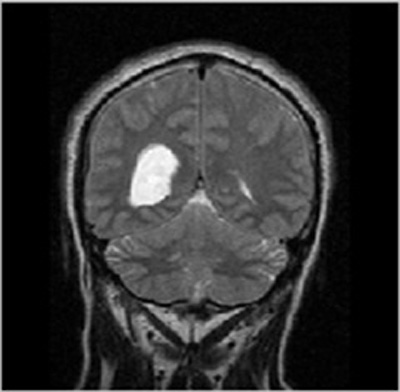
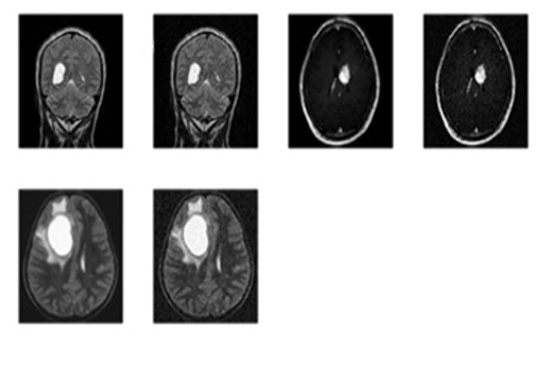
- Step-2: The MRI images were procured from BITE (Brain Images of Tumors for Evaluation database) by the NeuroImaging and Surgical Technologies Lab [7] as shown in Fig. (1).
- Step-3: Preprocessing of input MRI image [8].
- Step-4: The features were extricated from the preprocessed image for further processing. Each MRI image has its unique identification and the system would match various features like shape, color and intensity of the cancer cells with cells of image to declare whether it is cancerous or not. All these features were extracted using Scale Invariant Feature Transform and then the features were matched using HMM classifier [9]. 2 was chosen as the size of the erosion and dilation filter for morphological operations.
- Step-5: Hidden Markov Model (HMM) classifier was applied to segment cancerous portion in the MRI through 2D segmentation. HMM is an unsupervised model which is based on Markov Model according to which weights for generating output are the probabilities of sequence belonging to one category or the other depending on the output generated by the initial state [10]. It is based on posteriori probabilities that create a signed distance map from the mask after ensuring an image being a 2D matrix. It, then, determines the mean values and maintains CFL condition by evolving the matrix and calculating intermediate and final outputs with alpha being 0.2 and iterating the stages to produce correct results.
- Step-6: Image classification was done to differentiate cancerous and non-cancerous cells from the brain image using sorting factor kappa and properties of images that are shown through the bounding box.
- Step-7: Parameters such as accuracy, Peak Signal-to-Noise Ratio (PSNR), MSE (Mean Square Error) and Fault Rate Dust Detection were evaluated for selected image to know the accuracy of segmentation using implemented approach.
Accuracy= (TP+TN)/ (TP+TN+FP+FN)
Fault Rate Dust Detection= (TP+FN) / (TP+TN+FP+FN)
MSE= sum ((im1-im2). ^2)/ product (size (im1))
PSNR=10 log10 (256*256/MSE)
Where, TP- True Positive, TN- True Negative, FP- False Positive, FN- False Negative, im2- Filtered image
4. SOFTWARE USED
The software used for implementation of proposed work is MATLAB.
5. DESCRIPTION
The present study was conducted with the aim to detect tumor and classify soft and hard tissues in the image chosen from a set of T1- MRI images. The cancerous portion was extracted using HMM and it helped to classify the image into cancerous and non- cancerous portions. The entire work was implemented using MATLAB software. The entire task starting from giving input image to the classification of cancerous and non-cancerous tissues has been done following the stages of processes. A Graphical User Interface (GUI) was designed to enable the user choose the intended function. GUI is incorporated with the main code in which a push button is provided for intended task. The GUI is designed in such a way that the first button executes code to detect cancer from the selected image while the second button is used to calculate parameters for the same image. The GUI enables the user to select image from a set in which cancer has to be detected [11].
Firstly, the image to be considered for cancer detection is chosen from a window that pops up by clicking either of the two buttons- Cancer Detection or Parameter calculation. Then, the entire task of selecting an input image to the Classification and Segmentation using HMM is performed through different stages and the output will be classified tissues in MRI and segmented tumor portion. For the same image, parameters such as accuracy, fault rate dust detection, Mean Square Error (MSE) and Peak Signal-to-Noise Ratio (PSNR) were calculated to determine quality of detection.
The GUI specific for this application was designed to enable a layman to use the system for segmentation. A set of 5-6 images was selected from the database to implement the work. The images were attained by GUI and then, a window appeared to choose the desired image to check functionality of system. The selected MRI images were then converted into grayscale by resizing the image and filters were used to adjust intensity levels of image by removing noise. The MR image was given as input and that image was analyzed vertically and horizontally for the brain disorder detection. The line graph was plotted representing pixels analysis from various angles as per the vertical and horizontal analysis. Feature Extraction is generally employed to extricate the desired properties from the image depending on the application.
The features were extracted from the preprocessed image for further processing. Each MRI image has its unique identification and the system matches various features like shape, color and intensity of the cancer cells with cells of image to declare whether it is cancerous or not. Hidden Markov Model is employed in machine learning for classification and learning purposes to assign categories to parts of image on the basis of properties of maximum likelihood estimation by maximizing the expectancy of a pixel lying in a particular category.
Segmentation is a way of splitting an image into constituent segments. It is generally employed to identify features and other typical information within the image. In the presented work, the segmentation leads to the identification of brain disorder that aids in the process of medical diagnosis and treatment. In the absence of tumor, the system will give the same image as output with no marking while the presence of two tumors will give 2 marks in output image and so on. The technique of split and merge segmentation is used to segment the brain image on the basis of volumetric properties that partition the image on the basis of correlation and combine similar parts to distinguish the disease.
6. RESULTS AND DISCUSSION
To check functionality of the proposed work for the detection of brain cancer in MRI images using HMM classifier, a set of 6 images was obtained from BITE database. All images were processed to extract cancerous portion using HMM (Hidden Markov Model) classifier and then the corresponding parameters such as accuracy, Mean Square Error (MSE), Peak Signal- to- Noise Ratio (PSNR) and fault rate dust detection were calculated for detection using the HMM classifier. So when the code was run and an appropriate image was selected for the intended task of cancer detection and extraction of the abnormal portion, the first output was the figure showing input MRI image and the corresponding filtered image using subplot command to display them at position (1, 2, 1) and (1, 2, 2) respectively.
After the preprocessing stage, the image was scanned horizontally and vertically to locate the position of cancer and image features were extracted. The pixel properties were calculated and then compared to each of its neighbors to categorize each pixel into two classes- abnormal and normal cells in MRI input image. Hidden Markov Model [12] based classifier (HMM classifier) performed classification by distinguishing pixels into true and false classes by estimating the chances of occurrence of abnormal cells in the next event depending on the properties of each pixel. The larger the size of tumor, the faster the segmentation. Split and merge segmentation was also applied to detect tumor boundary.
In this way, a boundary box differentiates the regions of image with dissimilar properties in few seconds that further lead to the exact position marking of cancer pertaining in the brain and further extracts out the cancerous portion out of the MRI image fed as an input.
The parameters obtained using HMM were compared with other methods such as SVR Model that fed results of finite element simulations to machine learning algorithm, Neural Network model based on probability maximization, Genetic algorithm combined with fuzzy-C, transform based mathematical model that used similar parameters making it apt for comparison but all models employed different methodology.
The comparison shows that the implemented method has improved performance in terms of accuracy and error that highlights the merits of HMM over others through its efficient learning and good predictive properties but it includes intermediate stages too for computations.
The following figures show the output of Brain Cancer Detection. The image fed as input for brain cancer detection is shown in Figs. (1-7).
Table 1 shows the performance parameters- Peak Signal-to-Noise Ratio (PSNR), Mean Square Error (MSE), Fault Rate Dust Detection and Accuracy for six different images for cancer detection and classification using HMM.
The respective filtering output is shown in Fig. (2).
The average parametric values for HMM are shown in Table 2.
| S.No. | Feature | Significance of feature |
|---|---|---|
| 1. | Mean | Average value of net intensity of fed image. |
| 2. | Variance | Fluctuation of intensity change from average value. |
| 3. | Correlation | Signifies a value that associates every pixel to its nearby pixels transcending over whole image. |

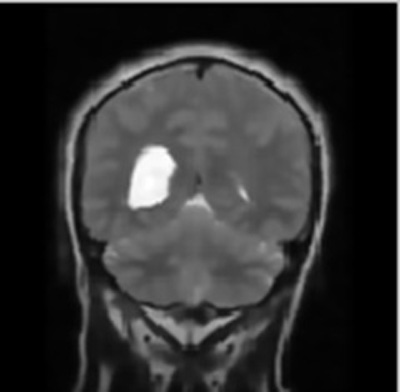
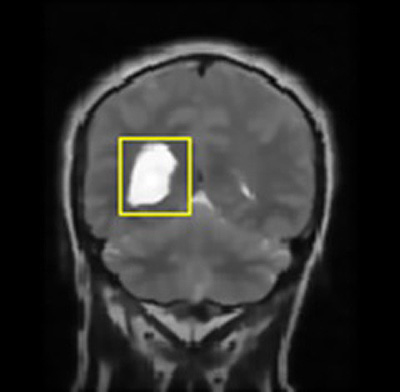
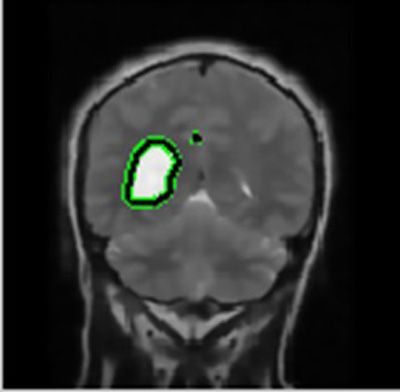

The same parameters namely- Peak Signal-to-Noise Ratio (PSNR), Mean Square Error (MSE), Fault Rate Dust Detection and Accuracy were obtained using the technique of Support Vector Regression (SVR) on the same set of images for brain cancer segmentation and classification for comparison with HMM.
The comparison of HMM and SVR was done in terms of Peak Signal-to-Noise Ratio (PSNR), Mean Square Error (MSE), Fault Rate Dust Detection and Accuracy shown in Table 3 given below.
The comparison of average Mean Square Error (MSE) achieved using HMM with Support Vector Regression (SVR) model [13] is shown in Table 4 given below.
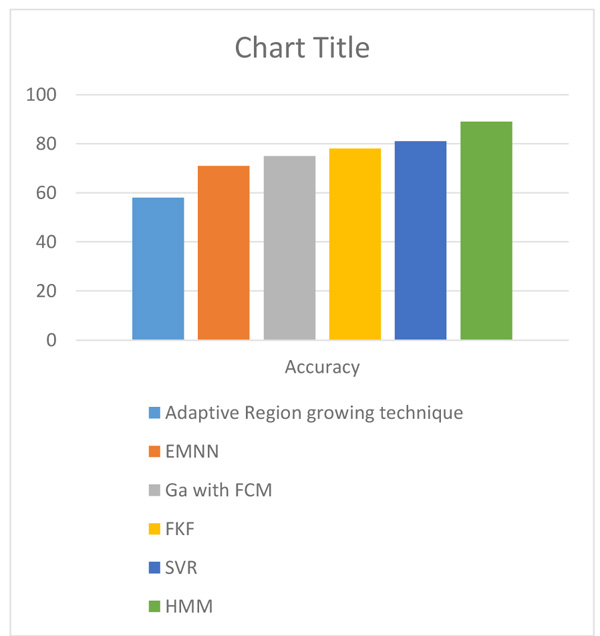
The comparison of average accuracy of different methods of brain cancer detection is shown in Table 5 below.
The comparison of average PSNR achieved using HMM was done with different methods of brain cancer detection from MRI images and it is shown in Tables 6 and 7.
| Images | PSNR | MSE | Fault Rate Dust Detection | Accuracy (%) |
|---|---|---|---|---|
| Image 1 | 20.55403 | 953.94374 | 0.64097 | 87.70700 |
| Image 2 | 20.73902 | 937.05068 | 0.87466 | 87.40067 |
| Image 3 | 19.88137 | 1057.94374 | 0.57028 | 89.03616 |
| Image 4 | 20.05673 | 1021.8977 | 0.81995 | 88.37820 |
| Image 5 | 22.40986 | 952.25156 | 0.48845 | 89.22137 |
| Image 6 | 23.90568 | 988.10963 | 0.92617 | 91.87735 |
| S. No. | Parameters | Values |
|---|---|---|
| 1. | PSNR | 21.257781 |
| 2. | MSE | 985.07166 |
| 3. | Fault rate dust detection | 0.72008 |
| 4. | Accuracy | 88.936% |
| S. No. | Parameters | SVR | HMM |
|---|---|---|---|
| 1. | PSNR | 16.984776 | 21.257781 |
| 2. | MSE | 1314.13322 | 985.07166 |
| 3. | Fault rate dust detection | 0.54001 | 0.72008 |
| 4. | Accuracy | 80.655% | 88.936% |
| S. No. | Parameters |
SVR Model (Tonutti et al., 2017) |
HMM (Proposed Method) |
|---|---|---|---|
| 1. | MSE | 1100.08 | 985.07166 |
| S. No. | Technique of detection | Accuracy |
|---|---|---|
| 1. | Adaptive region growing based technique (Sheela and Babu, 2016) |
58% |
| 2. | Expectation Maximum and Neural Network based Technique (EMNN) (Sheela and Babu, 2016) | 71% |
| 3. | GA with SVM (Gopal and Karnan, 2010) |
75% |
| 4. | Fuzzy bisector, K- means and F- PSO based technique (FKF) (Sheela and Babu, 2016) | 78% |
| 5. | SVR | 80.6% |
| 6. | HMM (Proposed Method) |
88.9% |
| S. No. | Technique of Detection | PSNR |
|---|---|---|
| 1. | K- means method (Mane et al., 2014) | 12.3986 |
| 2. | Watershed transform (Mane et al., 2014) | 11.2750 |
| 3. | Morphological operation (Borole and Kawathekar, 2016) | 14.4102 |
| 4. | SVR | 16.9847 |
| 5. | HMM (Proposed method) | 21.2577 |
The above shown tables summarize that the HMM method for brain cancer detection is better than others in terms of various parameters.
The improvement in accuracy of brain cancer detection in the proposed method over existing methods is shown in Fig. (8).
CONCLUSION AND FUTURE SCOPE
The implemented system assists in simple and quick detection of brain cancer in MRI images through a user interface designed for the ease of use. HMM classifier is used to classify cancerous and non-cancerous cells on the basis of properties of pixels through the process of scanning, segmentation and classification and detect cancer boundary leading to the extraction of cancer portion. The proposed method based on HMM has detected brain cancer with high accuracy nearly 88% with an improvement of around 8% over SVR and less errors resulting in high PSNR and better fault rate detection in comparison to SVR method. Also, the time taken for execution is less leading to quick detection by the system. So, it satisfies the need of developing fast and accurate methods to identify and extract cancerous portions from the input image automatically so that the problem can be curbed without disrupting body functionality permanently.
In future, the classification of different tissues for detection of cancer in medical images can be performed using other classifiers such as decision tree classifiers, naive bayes classifier etc. Moreover, the technique of raster scan can be employed for scanning. The recognition and extraction of various types of cancers like lung cancer, colon cancer etc. can also be done.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
The endeavor shall be incomplete without the acknowledgement to the almighty and all those personalities who have extended full co-operation and valuable assistance to us in making this research paper a reality. I would thus, like to take this opportunity to convey my deepest feelings of gratitude to all for giving this paper the final and desired shape. Our families, some supportive friends and the teachers have always been a support and source of great inspiration.


