All published articles of this journal are available on ScienceDirect.
A Comprehensive Study on the Application of Machine Learning Algorithms in the Prognosis of Ovarian Cancer
Abstract
Ovarian cancer is the third leading type of cancer found in women in India and ranks seventh globally. Several studies have shown that the population affected by ovarian cancer is profound to increase in the future. It is necessary to take steps for identifying cancer at the early stages to avoid mortality and recurrence. This chapter aims to survey the different ways machine learning models have been used in the prognosis of ovarian cancer - to predict the disease progression, recurrence, and mortality rate; analysis of genomic data sets; correlations and pattern analysis, and finding risk factors. The effective analytics on the imaging and other forms of data available from the patient’s electronic health records could unveil the possibilities of better or early diagnosis of ovarian cancer. The chapter will summarize the taxonomy of the various ways in which machine learning helps in ovarian cancer diagnosis, early detection, and treatment. In addition to surveying the current state-of-the-art application of machine learning algorithms for ovarian cancer diagnosis, the chapter aims to provide future research directions.
1. INTRODUCTION
The female reproductive tract contains two ovaries on the uterus side, which develops eggs for reproduction, and estrogens and progesterone production. Cancer that starts spreading in the ovaries is called ovarian cancer. Overgrowth of non-susceptible cells inside the ovaries leads to ovarian carcinoma. These cells may never die and accumulated to form a tumor. It is one of the most life-threatening cancers among women in the world. As per statistics, women aged 50-65 with high-risk factors are prone to develop ovarian cancer.
The Major key risk in the development of ovarian cancer are as follows (i) Family history of ovarian or colon cancer, (ii) Women in the period of Postmenopausal (iii) Genetic mutations. Compared to other gynaecological cancer, the survival rate of ovarian cancer patients is meagre. Ovarian carcinoma is coined as a “Silent killer” disease because there is no evidence of patients' symptoms until the last stage.
Cancer cells may travel to other parts of the body and multiply there, but the type of cancer is diagnosed with the name after finding its primary location. The state where cancer cells travel and start growing in some other part of inside the body is called metastasis [ 1]. Among all gynaecological cancer, ovarian cancer reports 2.5% of cancers in women.
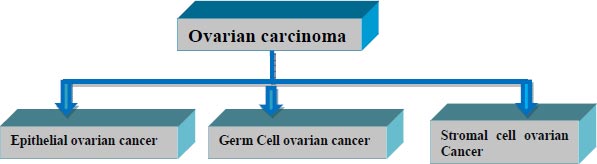
Based on the type of cell it originates from, ovarian cancer is categorized into three types, as shown in Fig. ( 1). More than 30 sorts of cancer arise from one of these three types.
The epithelial type of ovarian cancer conceives from the cell that covers the lateral surface of the ovaries. Over 90% of ovarian cancer belong to this category. Miserably Epithelial type doesn’t show any symptoms till the last stage of cancer.
Germ cell type of malignancy originates from the cells that produce eggs inside the ovary. These types of carcinomas generally affect youths and are easily treated. Over 5% of women are affected by Germ cell-type cancer.
Stromal cell ovarian carcinoma is a rare disorder that evolves from the connective tissues of ovaries. Nearly 5% of women are affected by this category.
Unlike Epithelial and germ cell types of cancer, the Stromal type has many symptoms such as abnormal hormonal production, menstrual cycle change, etc. [ 2].
| Stage Grouping | Description | Survival Rate (Approximately) | Therapy |
|---|---|---|---|
| Stage I | In this type of cancer, cells are cramped to ovaries and don't spread to any other ovary parts. | 90% | Abdominal hysterectomy, Removal of both ovaries and fallopian tubes |
| Stage II | In this cancer stage, cells may spread around the ovaries, parts such as fallopian tubes, and the uterus may get affected. | 70% | In addition to hysterectomy, specimens around the ovaries are tested for infection. |
| Stage III | The cancer cells may spread beyond the ovaries. It may start laying on the surface of the liver. | 39% | Treatment for this stage is the same as stage II. After surgery, the patient might get a chemotherapy procedure if needed. |
| Stage IV | Cancer has spread to all parts of the body. Fluids taken from other internal organs may contain cancerous cells. | 17% | Try to remove the parts where the tumor has spread with the combination of chemotherapy and Radiation therapy. |
The percentage of cancer spread inside the patient's body is found by a global system called staging. It helps in deciding the therapy for patients. The categorization of staging with its symptoms and causes is described in Table 1.
The disease can be diagnosed with vaginal ultrasound, CA125 blood test (Biomarkers, usually elevated in ovarian cancer patients), C.T. scans, and biopsy. Therapists generally perform a biopsy, a crucial test that confirms the presence of cancerous cells in ovaries.
Each stage of cancer has different kinds of symptoms. Usually Stage I cancer patient reports very mild or no symptoms at all. In the later stages, symptoms such as frequent urination, constipation, and bloating may occur [ 3, 4].
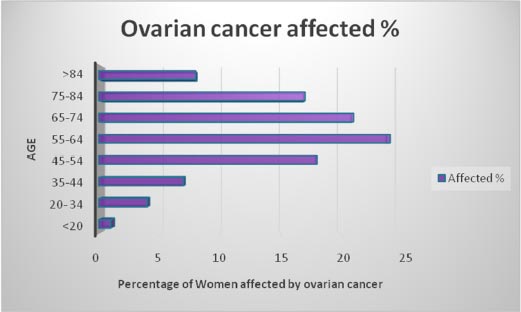
Earlier identification of ovarian cancer even before symptoms occur can increase the survival rate of the patients. Various data collected from patients can be stored in analytical sandboxes and analyzed further to predict this deadly disease. Fig. ( 2) shows the percentage of affected women concerning their age. Middle-aged women are most affected by this disease.
This survey aims at performing a comparison analysis between Machine learning, Deep Learning, and NLP Techniques, which uses various algorithms for the prediction. An accurate forecast for ovarian cancer must satisfy stringent data requirements. The data, such as the patient's profile, images taken from C.T. scan-Ray, and Gene expression is taken into account. Its required algorithm for analysis is intensely surveyed in this paper.
This review is divided into five sections: Section 2 expresses the Background of existing work in ovarian cancer prognosis. Section 3 briefly explains the Taxonomy of Ovarian cancer prognosis. In section 4, sources available for predicting ovarian cancer are briefly narrated. The various techniques for processing the data are outlined in section 5. The final section concludes the survey with future directions.
2. BACKGROUND
Cells are an essential element for all living beings, responsible for providing energy to the body. Each cell in the human body is comprised of Deoxyribonucleic acid (DNA).DNA contains four types of chemical bases Adenine (A), Guanine (G), Cytosine (C), and Thymine (T) that form the sequence arrangement for building any living creature. Genes are made from Deoxyribonucleic acid (DNA). Each cell in the Human body is wrapped with a thread-like DNA structure called chromosomes. Flaws may occur when the section divides them, which leads to mutation. In addition to environmental factors, sometimes humans carry these flawed genes from their parents. The cell which drastically divides themselves neither repair nor dies will lead to a deadly disease called cancer. Gene expression patterns reflect the discrepancies in the survival of advanced ovarian carcinoma.
2.1. Machine Learning in Cancer Prognosis
Identifying ovarian cancer in the early stages is challenging because it doesn't show unusual symptoms in its initial phase. The diagnostic study for various types of ovarian cancer is identified with different kinds of machine learning algorithms Various classification and clustering algorithms in Machine learning are used in identifying ovarian carcinoma in its early stage. In cancer diagnosis, the focal point is: 1) Predicting the survivability of cancer patients 2) Predicting the reoccurrence of various types of cancer.
2.1.1. Feature Selection Process and Machine Learning Models
The process of feature selection involves automatically choosing the best feature that contributes more to the predictor variable of machine learning models. The feature selection method (FS) aids in improving machine learning model accuracy by decreasing overfitting and simplifies the machine learning model easier to interpret.
The major types of feature selection methods are filter methods, wrapper methods, and embedded methods.
2.1.1.1. Filter Methods
Filter methods capture the unique features assessed by univariate statistics. It performs well with high-dimensional data. Compared to wrapper methods, these techniques are quicker and more computationally efficient.
2.1.1.2. Wrapper Methods
By comparing every possible feature combination to the evaluation criterion, wrapper methods implement a greedy search methodology. It gives better accuracy than filter methods.
2.1.1.3. Embedded Methods
Embedded methods are a combination of filter and wrapper methods.
The authors of a study] used a cervical cancer dataset from UCI Machine Learning Repository collected from Hospital Universitario de Caracas in Caracas, Venezuela [ 5 ] This dataset contains the medicinal histories of 858 patients with 36 attributes. By applying Support Vector Machine, K Nearest Neighbor, Random Forest, and C5.0 authors predicted the occurrence of cervical cancer.
Prediction of Ovarian cancer (O.C.) is an essential process in the medical field. The authors suggested that the prognosis of the tumor is in the category of benign or malignant cancer. Cancer value is used to predict the stage of cancer accurately. In one of the papers,a total of 349 patient details were analyzed with the features of demographics, blood routine tests, general chemistry, and tumor markers [ 6]. In total, 349 attributes were analyzed in this test. Out of 349 patients, 235 patient records were tested. The essential prediction features were extracted using the Machine learning Minimum Redundancy – Maximum Relevance (MRMR) method. Out of which human epididymis protein four and carcinoembryonic antigen were considered to be the most important feature. It has been compared with the Relief F method for a better feature extraction process. One of the required machine learning classification methods, logistic regression, was used to classify the data. Besides, the decision tree and risk of ovarian malignancy algorithm have been compared for better classification. Based on the benignant tumor size, the data was classified in a decision tree. Benign ovarian tumors and ovarian cancers are easily classified with machine learning approaches [ 6].
Laios et al. used the Chi Square test of independence and the Minimum redundancy maximum relevance feature selection method to extract the best features from the categorical data. Various machine learning models including conventional regression analysis were compared For HGSOC prognosis [ 7].
2.2. Deep Learning in Cancer Prognosis
Deep Learning is a subset of Artificial intelligence that imitates the human brain structure, which helps us decide with different kinds of data such as text, images, and genomic data. The accuracy of results plays a vital role in ovarian cancer survivability and early prediction. Regarding the voluminous data, deep learning models provide better accuracy with fewer efforts. The classification of neural networks is based on the type of data we are giving as input. The text type of data is handled by Recurrent Neural Network (RNN). The classification of image data is performed by Convolution neural networks (CNN) [ 8].
The image dataset collected from Guru Gobind Singh Medical College and Hospital Faridkot contained a benign and malignant tumor. The CT images had a record of different stages of ovarian cancer. The initial step began with the pre-processing of raw images. Fuzzy c means the algorithm was used to identify specific regions in the pictures. Various features of the images were retrieved from the scale-invariant feature transform algorithm. Optimized results were obtained using a genetic algorithm. Finally, CNN based classifier was developed for the prediction of various types of cancer [ 9 ].
The RNAseq dataset for Lung, Breast, and stomach cancer was collected from TCGA. To avoid the same gene expression data DeSeq method was being used. The neural network was framed with multiple layers to find the optimal prediction [ 10].
Based on genomic variations, 12 different types of cancer can be predicted using deep neural networks. A deep neural network is built within the Tensor flow model. The Genomic Deep Learning (GDL) method identifies mutation that transforms a normal cell into cancer cells. This GDL identifies cancer in its initial phase [ 11].
Padideh Danaee et al. proposed an in-depth learning approach using a stacked Denoising Autoencoder to extract the relevant feature from gene expression data. Comparison has been made for appropriate feature extraction using principal component analysis and Kernal Principal Component Analysis (KPCA). Further classification of genes related to breast cancer was done using SVM-RBF and deep neural networks [ 12].
2.3. Natural Language Processing in Cancer Prognosis
Natural Language Processing (NLP) plays a crucial role in making effective decisions in the healthcare industry. It helps in extracting meaningful features from vocal or scripted documents. In cancer treatment, NLP is used for classification, disease recurrence, and various cancer stages. The details collected from the annotated corpus are used for predicting the staging of cancer. After extracting the relevant information, the relation between the entities is made [ 13].
Cancer-associated information is retrieved from various sources such as scientific papers, health records, and annotated databases. NLP's first step is to categorize named entity recognition by assigning the word to a predefined category. The next step is information extraction, which automatically extracts the structured data from unorganized data. With the best text classification algorithm's help, the information received is placed under a systematic group. The final step is to retrieve the required information from the health records and other medical-related data repositories [ 14].
3. DATA PROCESSING TECHNIQUES
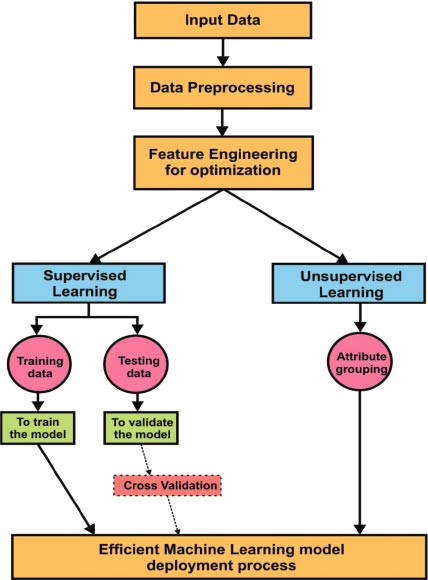
Fig. ( 3) explains Machine Learning approaches in detail. Input data is received through various sources. Later data pre-processing is done for better optimization through data engineering techniques. In supervised learning, data was split into two parts: Training data and testing data. Training data is used to train the model to obtain better accuracy. Test data is used to validate the model. Validation can be done using various methods. Cross-validation is one of the best techniques during the verification of the model. The model is validated with a small sample of values in the given data set is called cross-validation. In unsupervised learning, the attribute grouping process was implemented for grouping similar kind of features. Dimensionality reduction is also an efficient process in feature engineering. It helps to achieve improved accuracy with less number of attributes. Finally, the efficient machine learning model is used in the deployment of applications.
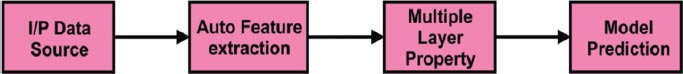
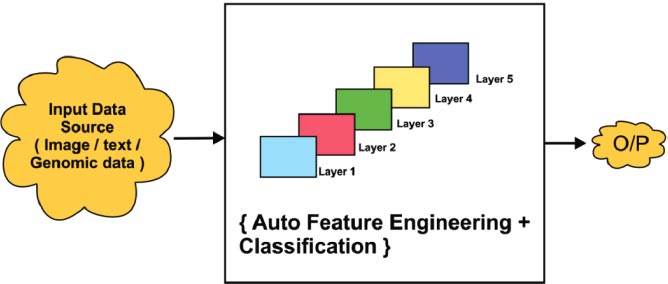
In Fig. ( 4), the General flow of the Deep learning process is depicted. Deep learning has the essential characteristics to obtain better accuracy is called automatic feature extraction. Also, it has the multilayer property to do in-depth analytics. Finally, the accuracy of the model prediction is done using improved and deep training techniques.
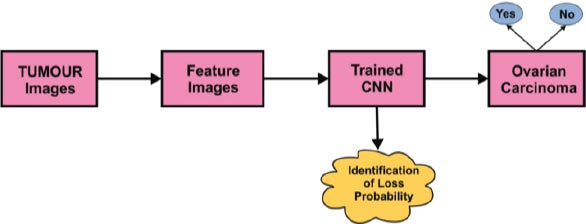
The in-depth learning image classification process using the Convolution Neural Network (CNN) algorithm is depicted in Fig. ( 5). With sample tumor images as a reference, the relevant image feature identification process has to be completed. After features were retrieved, the data was fed to the Convolution Neural Network classifier. The trained CNN will predict whether the given input contains traces of ovarian carcinoma or not.
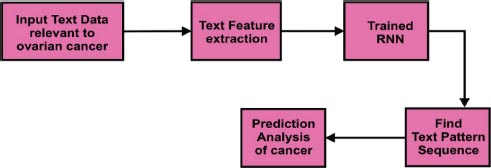
In Fig. ( 6), the text classification process was explained using an in-depth learning approach. Text data relevant to ovarian cancer was considered as input. Relevant features were extracted using text feature extraction methods. The Recurrent Neural Network (RNN) was used to identify the next word's sequence in the text data. It would be helpful to do the prediction analysis of cancer efficiently.
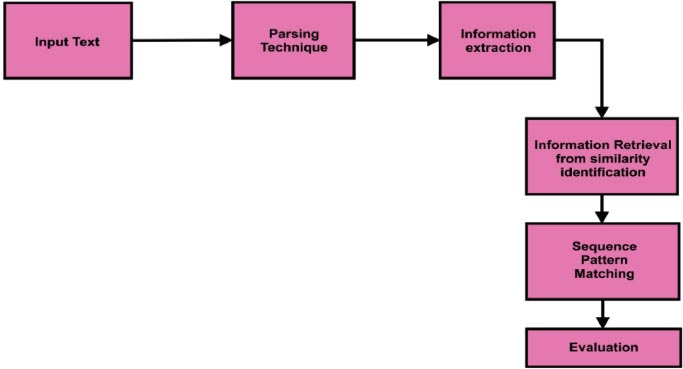
Fig. ( 7) depicts the workflow of natural language processing. Input data was split using the parsing technique. The information extraction process was implemented after parsing the input text. Similar word identification and sequence pattern patching were implemented during the Information retrieval process. Useful text classification was evaluated after the information retrieval process.
The general architecture of the deep learning process with multiple hidden layers is shown in Fig. ( 8). Deep learning always provides better accuracy due to the multilayer protocol. Input data is collected from various sources such as image, text, or genomic representation. It will be classified using multiple layers. In detail, general learning consists of multiple hidden layers for analyzing the data in depth. Fig. ( 6) shows that there are five layers that act as hidden layers. In that hidden layer, the auto feature extraction, and classification process was implemented efficiently. Finally, the output was obtained with improved classification accuracy.
4. TAXONOMY OF OVARIAN CANCER PROGNOSIS
Ovarian cancer prognosis can be done in three ways: image analytics, text analytics, or Genomic data analysis. The data was received from various sources, and then it was pre-processed in the data staging technique. Pre-processing of information is very important for any kind of analytics. Null values, irrelevant features, and missing values always reduce model prediction accuracy. After the pre-processing of data, it was analyzed using various machine learning algorithms. Machine learning, Deep Learning, and Natural Language Processing are considered in our taxonomy. In Machine Learn- ing, two major categories of the learning environment were considered, supervised, and unsupervised learning (Fig. 9).
In supervised learning, the classification of data is handled by various classification algorithms like Naive Bayes, decision tree, support vector machine, K nearest neighbour (KNN), and logistic regression. An efficient clustering process is done in the unsupervised environment by various clustering algorithms like k means, DBSCAN (Density-Based Spatial Clustering of Applications with Noise), Gaussian mixture, and hierarchical clustering model. In the Deep learning model, the classification process is handled by various algorithms like Recurrent Neural Networks (RNN), Convolution Neural Networks (CNN), Autoencoder, Deep Belief Networks (DBF), and Radial Basis Function, etc.
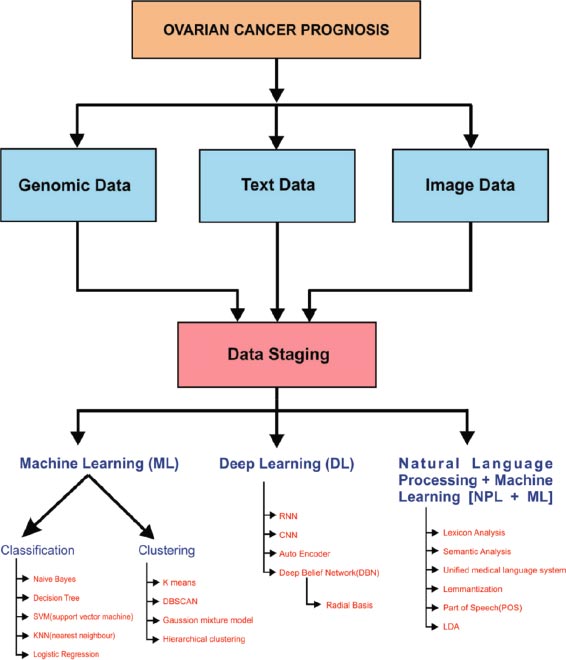
Natural language processing handles classification by Lexicon approaches semantic analysis, Unified Medical Language system, speech, and lemmatization. The correlation between gene expression and phenotype data such as tumor morphology, a consequence of chemotherapy, and a related blood test from the laboratory should be studied. NLP is used to extract cancer-related information automatically from electronic medical records.
5. VARIETY OF SOURCES: TEXT, IMAGES, AND GENOMIC DATA
The sources include personal health data, and CT images; MRI images are used in the prediction and recurrence analysis of ovarian cancer. Ovarian cancer can be detected with ultrasound, high CA125 values, Positron emission tomography (PET), and biopsy. The sources of images are helpful in the detection, portrayal, and detecting the stages of ovarian cancer. The benign and malignant tumor is easily distinguished with image sources. Various types of ovarian carcinoma, such as Epithelial, Stromal, and Germ cells are easily detected with the radiologic appearance of Topographic images. The mutation in a specific type of gene is a primary reason for causing ovarian cancer. Data related to gene mutations and personal health data can be integrated to predict cancer early.
An ensemble approach for the prognosis of ovarian cancer is developed by Kawakami et al. [ 15]. Based on the blood biomarkers dataset. Clinical parameters based on ovarian carcinoma and ovarian tumors are differentiated using EOC blood markers. Multiple logistic regression analysis is made to choose the numerous variables as predictors in bio-blood markers. With the help of machine learning approaches, risky patients are identified prior and preliminary treatment for them can be provided.
An ensemble-based approach is proposed by Tarek et al. [ 16] for the classification of different types of gene data associated with various types of cancer. Each ensemble approach chooses the best feature selection method for improvising the accuracy in cancer classification.
A study on Genetic profiles with methylation data plays a vital role in the prediction of ovarian cancer. Methylation is a procedure that includes methyl groups in the existing DNA sequence. Main feature selection methods are used to select the useful feature from the combination of gene and methylation profiles. Gene profiles collected from the TCGA repository and methylation beta values play a vital role in ovarian cancer [ 17].
Li et al. developed an integrated framework that predicts recovery chances in the chemotherapy process. The support vector machine algorithm is applied as a base model to classify the data with necessary profile information and merge it with gene expression. The Leave-One-Out Cross Validation method was used to evaluate the performance measure of both models. The final results showed that the fusion of clinical information and gene expression data provided better accuracy [ 18].
Hybrid machine learning algorithms such as SVM, C5.0, MARS (Multivariate Adaptive Regression Spines), and Random forest were implemented over the patient's profile to predict persistent ovarian cancer [ 19].
Certain types of genes are responsible for causing the deadly disease of ovarian cancer. The mutations in BRCA1, BRCA2, and TP53 Genes are linked to causing hereditary ovarian cancer. Screening based on gene expression prevents this type of hereditary cancer. Genetic testing can be done by taking the sample from blood, saliva, hair, tissue, etc.; integration of Genetic based testing in ovarian cancer reduces the risk and identification of this disease in its early stage. It is estimated that 64% of women will develop ovarian cancer by BRCA1 and BRCA2 gene mutations [ 20].
Ahn et al. performs prediction of ovarian cancer based on an RNAseq dataset collected from The Cancer Genome Atlas (TCGA). Genes were marked as 'good' and 'bad' based on Genes coefficient calculation. The study shows the classification of high-risk and low-risk of patients [ 21].
With the K-means clustering technique's help, the ovarian cancer Gene dataset from TCGA (The Cancer Genome Atlas) is subdivided into three types. The Dunn index metric is used to evaluate the performance of the clusters. Based on the survival rate of the cancer, patient’s morphological type is defined. The ensemble classifier predicts each subtype. The endurance of patients in all three subtypes is significantly different risks. They used two different data sets for their experimental purpose. One was from TCGA, and the is an independent dataset collected from previous work Gyorffy et al., 2012 [ 22]. They combined and utilized the drug target information from the following three different types of databases. They were the Therapeutic Target Database, Drug Bank, and Drug-Gene Interaction Database. Using the Pearson correlation coefficient and rank-based method, they constructed each subtype's co-expression network. They compared their work with two different controls. The first control applied a t-test to select the features and the second control used Euclidean distance of the vector to assign the patient with a similar subtype. Subsequently, they predicted the prognosis. They have done a survival analysis of cancer patients in all three types. The resultant model showed better significance than the existing methods in classifying the patients in different survival risks [ 23].
The authors in a study identified diffusion-weighted image value using an MRI scan method [ 24]. Epithelial ovarian cancer patient tumor load is calculated and can be analyzed whether the complete cytoreduction is achieved. In this paper, the peritoneal cancer index value has been compared in 2 stages. Stage one is the MRI scan period and step 2 is surgery time. The entire cytoreduction is evaluated in both locations using ROC. The correlation coefficient has been identified in both phases. As a result, it was observed that the stage of MRI provides better accuracy than the surgery time to predict the cytoreduction of ovarian cancer.
Gopichand et al. clarifies in real-time investigation predicting patient response to chemotherapy for ovarian cancer is analyzed using C.T. scan images. The patient response was calculated according to the shape, density, texture, and wavelet of the image feature. At the time of wavelet transmission was categorized in 4 categories Low-Low, Low –high, high –low, and high-high. (L-H: applying short scale to high scale filter). The authors have identified the feature selection process in two different stages: pre-treatment and post-treatment. In the pre-treatment stage, some of the image features are recognized as the best features. They are skewness, contrast, uniformity, entropy, density, kurtosis, and compactness. In the post-treatment stage, in addition to the characteristics of pre statement stage, some new features were also considered like surface area, energy, media, volume, etc. [ 25].
The recurrence of ovarian cancer causes significant risk in the survival of cancer patients [ 26]. Wang et al. proposed an unsupervised deep learning method for extracting required features from the CT images. The raw images are converted into 16-dimensional features. The decoder is used to reconstruct the original image of the tumor. The radiologist has been used to identify the tumor part from the CT images. The Cox proportional-hazards regression model is used to find the recurrence of cancer in individual patients. The feature selected is combined with personal clinical characteristics for better predictions [ 27].
The following Table 2 summarizes the various models and their accuracy for predicting ovarian carcinoma.
| References | Model | Cancer Type | Type of Data | Validation Method |
|---|---|---|---|---|
| [ 6] | Machine learning Minimum Redundancy - Maximum Relevance (MRMR) feature selection method & Decision tree | Ovarian cancer | Demographics, blood routine test, general chemistry, and tumour markers dataset. | Compared with the risk of ovarian malignancy algorithm (ROMA) and logistic regression model. |
| [ 8] | Feature Extraction--Scale-invariant feature transform algorithm
Convolution Neural network |
Stages in ovarian cancer | Ovarian cancer image | Genetic algorithm |
| [ 11] | Deep Neural network | Breast cancer, colon cancer, glioblastoma multiform, kidney renal clear cell cancer, low-grade glioma, lung squamous cell cancer, Ovarian cancer, prostate cancer, Skin cutaneous melanoma, Thyroid cancer, and Uterine corpus endometrial cancer. | Gene mutation data | Sensitivity and specificity formula to evaluate the classifiers’ performance. |
| [ 15] | 1. Gradient Boosting Machine
2. Support Vector Machine 3. Random Forest 4. Conditional Random Forest 5. Naive Bayes 6. Neural Network 7. Elastic Net |
Epithelial ovarian cancer | Blood biomarkers | 10-fold cross-validation |
| [ 17] | 1. Supervised Principal component and the 2.Elastic net method | Ovarian cancer | Gene expression and methylation profiles | 2-fold cross validation. |
| [ 18] | Support vector machine | Chemo-response for ovarian cancer | Gene expression data | Leave-one-out cross-validation |
| [ 21] | Univariate Cox PH model | LASSO | Microarray and RNASeq data | N/A |
| [ 23] | Clustering analysis based on subtypes | Ovarian cancer | Genomic | Leave-one-out and independent validation |
| [ 24] | MRI PCI and diffusion-weighted imaging | Epithelial ovarian cancer | MRI Image data | Receiver Operating Characteristic (ROC) Curve |
| [ 25] | Equal-weighted fusion method and CAD | Ovarian cancer | Computed tomography (CT) images | RECIST |
| References | Model | Type of Data | Accuracy | Significant Algorithm Settings Discussed |
|---|---|---|---|---|
| [ 28] | Deep interactive learning with a pretrained segmentation model | Image | 86 | Stochastic gradient descent |
| [ 29] | Convolutional Neural networks | MRI sequence | 81 | CNN with Xception architecture |
| [ 30] | Hybrid evolutionary deep learning model, using multi-modal data | Gene expression | 98.8 | Ant Lion Optimizer |
| [ 31] | Supervised Deep learning approaches | Histopathology images | 88.2 | 5-fold cross-validation |
| [ 32] | Cox Proportional Hazards Model | Histopathological images | 88.2 | - |
| [ 33] | Deep convolutional neural network | Image | 86.9 | DenseNet-121 |
| [ 34] | DCNN | Images | 81.38 | - |
| [ 35] | Deep Convolutional Neural Networks (DCNN) | Histopathological images. | 84.64 | Visual Geometry Group – 16 |
| [ 36] | Variational Autoencoder | Gene Expression | 81.44 | Min-max, SMOTE, ReLU activation function |
| [ 37] | Multi-layer perceptron and Logistic Regression | Numerical | 99 | PCA, Stochastic Gradient |
| [ 38] | Improved RCNN | Images | 93.2 | Attention-based activation function |
| [ 39] | GoogleNet+Random Forest Classifier | Ultrasound images | 99.79 | Softmax activation function, Reverse conduction optimization |
| [ 40] | Residual Encoder Decoder CNN | CT images | 95.2 | Adam optimizer |
In addition to the above work related to applying machine learning algorithms, the recent work in the prognosis of ovarian carcinoma involving deep learning techniques is summarized below. The class of deep learning algorithms has considerable advantages - automating feature extraction, efficiency of understanding unstructured data, appreciable self-improvement capability with feedback, and ease of parallelizing to name a few. Deep learning algorithms use different layers of neural networks allowing them to learn from huge volumes of data. The role of deep learning in diagnosing ovarian cancer is summarized in Table 3.
CONCLUSION AND RESEARCH DIRECTIONS
This review endeavored to elucidate and assess the performance of various machine learning and deep learning methods in the prediction of ovarian cancer. The taxonomy for ovarian cancer prognosis is presented concerning text, image, and genomic data. In particular, we compared the pattern observed concerning the type of model being used, the nature of cancer diagnosed, the kind of data incorporated in the prediction and overall accuracy of the various methods with its validation techniques, and the effective analytics in ovarian cancer prediction. The survey emphasized a strong bias towards Machine learning, deep learning, and NLP models in the earlier forecast of this deadly disease. In this review, we outlined the performance of deep learning models with similar or improved accuracy as compared to machine learning models. Future research should extend by optimizing the existing algorithm and diagnosing ovarian carcinoma in its early stage.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflicts of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.


