All published articles of this journal are available on ScienceDirect.
Characterization of Cells Interactions with Patterned Azopolymer-Based Materials using SEM, AFM and Video Microscopy
Abstract
Introduction:
Artificial Extracellular Matrices (ECMs) are promising tools for the study of cell behaviors.
Methods:
Here, we report a protocol for the use of a reconfigurable biocompatible azopolymer thin film through a photoinduced reconfigurable structuration of its surface to study nerve growth, differentiation and cell guidance.
Results & Discussion:
We show that this protocol combined with a spontaneous self-photoinduced polymer is suitable for time-lapse fluorescence video microscopy and can be easily adapted to electron microscopy techniques (SEM) and near-field imaging techniques (AFM).
1. INTRODUCTION
Since several years, numerous works and experiments show that cells develop a high sensitivity for chemical and physical features of 2D and 3D substrates used in the goal to control and study their in-vitro proliferation. In this view it has been shown that several components of the artificial Extracellular Matrices (ECM) affect different comportments of cells during growths such as adhesion, proliferation, differentiation, and cell death [1-3]. Several signals induced by the material substrate, such as 2D and 3D organization, influence cell’s comportments and thus highlighted the paramount importance of ECM. Topographic cues are known to induce and influence cell’s growth and functions. Different methods and techniques were developed to produce micro- and nano-grooved materials. The final objectives are aimed to study contact interactions, guidance and mechano-transduction phenomena [2, 4-6].
The development, inscription, and modulation of permanent patterns on several substrates usually associate microfabrication and nanofabrication methods such as soft lithography, electron or focused ion beam lithography [4-7]. One of the advantages of the latter is the very high spatial resolution given by the energy used to inscribe a topographical pattern. However, these techniques often require expensive equipment, specialized technicians and are time consuming, especially when large surfaces are processed. Potentially additional techniques, such as embossing, can be used for large production of samples [8]. Moreover, once manufactured, the geometric, topographical and mechanical features of the substrate surface are often fixed and thus cannot be easily modified afterward. Finally, the designed topography being intrinsically robust to any changes and modifications which is, basically, in contrast with in-vivo ECM [9].
To overcome these limits and create more versatile and adaptive biological substrates for dynamic time lapse cell investigations, the use of stimulus-responsive materials as biological supports could reflect in-vivo situations [4]. These recent works used azopolymer-based materials as substrates in which patterns are remotely transferred on its surface using light properties. These holographic photo-printing methods of Surface Relief Gratings (SRGs) on azopolymer films is a promising approach for a straightforward and easily reconfigurable fabrication of biological substrates even after cells adhesions [7]. A linearly polarized light is used to create tight photo-induced and spatially controlled gratings, like a holographic pattern. A circularly polarized incoherent light or temperature above the Tg allows the possibility to modify the pattern and to totally or partially erase it.
An accurate imaging of the photoinduced surface of the samples using this new azopolymer-based material is mandatory to assess its architecture, before and after incubation in cell culture media, but also to precise the morphology and development of seeded cells, and the intimate cells/substrate relationship. Scanning Electron Microscopy (SEM) has been used for many years to characterize nanotechnologies and biological samples at the nanometer scale [7]. SEM allows a high-resolution imaging with a large depth of field and can be combined with other light microscopy techniques such as confocal microscopy. However, one of the challenges using SEM imaging is to use a preparation procedure that does not induce any morphological alteration. Moreover, sample preparation should not also modify the structure of the polymer substrate. However, photochromic and photoinduced patterns are often sensitive to chemicals used in the dehydration step compulsory for SEM, and as such may lead to preparation artefacts. Therefore, the ability to view hydrated, non-conductive samples without treatment, destruction or semi-destruction requires the use of SEM a difficult technique to view biological samples in particular when the surface samples are made with organic photostructured polymer-based materials [3, 7].
In this study, we aimed to establish a protocol to allow the use of SEM for a high-resolution imaging of patterned azopolymer materials before and after seeded cells, with a particular reference to possible sources of damages during the preparation of the samples. We also emphasize that this protocol can be used for anti-bacterial cleaning for any photostructured surface polymer or more generally to any polymeric surfaces.
2. EXPERIMENTAL SECTION
2.1. Azolymer
Films with a thickness of 170 μm (Fisherscientific) were prepared on coverslips from an azopolymer solution using a spin-coating technique. Polymer films were made with a highly photoactive azobenzene derivative containing heterocyclic sulfonamide moieties with a huge capacity for surface patterning. The details of chemical synthesis of 3-[{4-[(E)-(4-{[(2,6-dimethyl- pyrimidin-4-yl) amino] sulfonyl} phenyl) diazenyl] phenyl} - (methyl) amino] propyl2- methylacrylate have been previously reported [10]. The synthesized polymer was dissolved in THF (40 mg/1.2 ml), ultrasonicated for 30 min, filtered through a syringe filter and spin-coated at 1200 rpm for 40 s on a pre-cleaned glass coverslip. After films deposition, the samples were put in an oven at 50 °C during 24h for drying and removing of any residual solvent. The film thickness, determined by a Dektak Profilometer, was around 550 - 600 nm. The molecular mass of the polymer determined by GPC was between 14000 and 19000. The glass transition temperature was estimated at 57.5 °C.
2.2. Surface Relief Grating Inscription
A diode pumped solid state laser, operating at λ= 473 nm, was used to excite the azopolymer close to its maximum absorption wavelength. The sample was placed perpendicular to the direction of the laser beam and irradiated by a beam with a diameter size of 3 mm at 1/e2 and a power density of 0.7 W/cm2. The inscription of the surface pattern only needed a single beam, the pattern being produced by a molecular self-organization of the material during the laser illumination by a trans-cis photoisomerization. It resulted in a locally preferred orientation of the azobenzene groups, oriented perpendicularly to the incident electrical field of the laser. A polymer mass migration occurred, inducing a pattern inscription on the material surface. The sample was illuminated during 15 minutes, until the complete structuration of the thin film surface, which was determined by the saturation of the diffraction intensity. The pattern pitch was calculated using the formula 2d = λ/.sin(Θ), where λ is the laser wavelength and Θ is the angle between the incident beam and the mirror. Patterns with different pitches could be prepared by varying the incident angle (Θ) of the laser.
2.3. Cell Culture
PC12 cells were maintained in DMEM medium (Dulbecco's Modified Eagle Medium, ThermoFisher Scientific, France) with 10% fetal calf serum (Biomedia, France) supplemented with 1% Non-Essential Amino-Acids (NEAA, Invitrogen, France) and antibiotics (1% Penicillin and Streptomycin; Invitrogen, France) [10] for one week to prevent any bacterial contamination. The medium was changed twice a week and cells were passed at 80% confluence. PC12 cells were transfected with pDsRed2-mito plasmid (Clontech®, France) using Viromer Red® (Lipocalyx, France) following the recommendation of the manufacturer. Cells were then treated 48h later with Gentamicin (G418, 350µg/ml; Sigma, France) and clones stably expressing pDsRed2 were selected, expanded and frozen for further experiments.
2.4. Time-Lapse Video Microscopy
PC12 stably expressing pDsRed2-mito were seeded at low density on coverslips previously coated with the polymer and treated for one week with Nerve Growth Factor (NGF; 100ng/ml, Sigma, France) as previously described [10]. Cells were then visualized using an inverted Leica microscope under bright field and fluorescence conditions in a thermostatic chamber DMI 6000B. Measurements were performed at a temperature of 37 °C. A Carl Zeiss (Jena, Germany) plan achromatic 10x oil immersion objective (0.25 NA) was used to enable a broad field of view. Images were taken every 60 seconds for 30 minutes using the Metamorph software (Microscopy Automation and Image Analysis software - Molecular devices). A phase contrast imaging was performed to analyze cell shapes and areas. The Neurite Outgrowth Applications Module allowed to measure and characterize outgrowths from the cell bodies. Acquisition of fluorescence images was performed to analyze pDsRed2-mito labeled mitochondria. Images were deconvolved using Huygens Essential® software (Scientific Volume Imaging, Hilversum, The Netherlands) and Imaris 8.0® software (Bitplane, Zurich, Switzerland) was used for 3D processing, morphometric analysis and mitochondria tracking.
2.5. Atomic Force Microscopy
Atomic Force Microscopy (AFM) measurements were performed in the tapping mode using a commercial AFM (Veeco, Autoprobe CP Research). The scan rate was 0.5 line/s and the oscillation frequency was approximatively 300 kHz. Data were processed and analyzed with the WSxM 5.0 software (Nanotec Electronics, Madrid, Spain).
2.6. Scanning Electron Microscopy
For electron microscopy characterization, 100 000 PC-12 cells were seeded on polymer-coated glass slides (Thermofischer, France). Cells were then treated with NGF for one week as previously described [10]. Coverslips were then rinsed in phosphate buffer (0.1M pH 7.4) and fixed in glutaraldehyde (4% in phosphate buffer) for 16 hours and rinsed three times in phosphate buffer. Samples were then post-fixed in osmium tetroxide (1% in water) for 60 min and rinsed three times in distilled water.
For dehydration, we could not use ethanol or butanol as they solubilized the polymer and damaged its surface. On the contrary, it was resistant to 2-propanol, as a substitute for ethanol, and to hexamethyldisilazane (HMDS). So, samples were dehydrated in three non-diluted 2-propanol baths during 30 min, then in a 2-propanol / hexamethyldisilazane (v/v) mixture bath during 45 min followed by two baths of HMDS, the first one during 45 min and the last one left until its complete evaporation. Then, in order to visualize cells, the samples were coated with a very thin layer of platinum with a thickness of 20-nm using a Leica EM ACE 600 (Leica, Rueil-Malmaison, France). Finally, samples were examined with a field emission scanning electron microscope (JEOL 6301 F, Paris, France) with an accelerating voltage of 3 kV under a vacuum of ≈ 10-5 mbars.
3. RESULTS
3.1. Azopolymer: An Adequate Material for Live Cells Imaging
PC12 cells transfected to express the DsRed2 fluorochrome reporter under a mitochondrial targeting signal were seeded on coverslips previously spin-coated with the new biocompatible azopolymer. As shown in the Fig. (1) cell bodies and neurites of transfected PC12 cells were easily observed after 5 days both using phase contrast and fluorescence imaging (Fig. 1A and B). Interestingly, neurite extensions could be well observed. They took the direction of the grating grooves. Arrowheads on Fig. (1C and D) show a neurite end (growth cone), developing toward the groove of the thin film surface pattern. The elongation of the tip of neurites (growth cone) was also visualized, associated with mitochondria trafficking (arrows on Fig. 1E and F). The cellular mechanical response to the photo-structured surfaces had an amplitude modulated surface with a typical pitch of 800 nm and a depth of 100 nm. Probably, as it was previously demonstrated [11], that the sensing apparatus of cellular rigidity can also detect discrete sub-micrometer discrepancies in the polymer matrix rigidity. The amplitude modulation of the azopolymer surface is the place for local modulation of the Young modulus due to the photo-induced modification of the material mechanical properties during the trans-cis isomerization [12, 13]. One possible suggestion is that this very small local modification takes place at the minimum of the grooves suggested by a modification of the Raman signal [12, 13].

3.2. Azopolymer Use for Time-Lapse Imaging
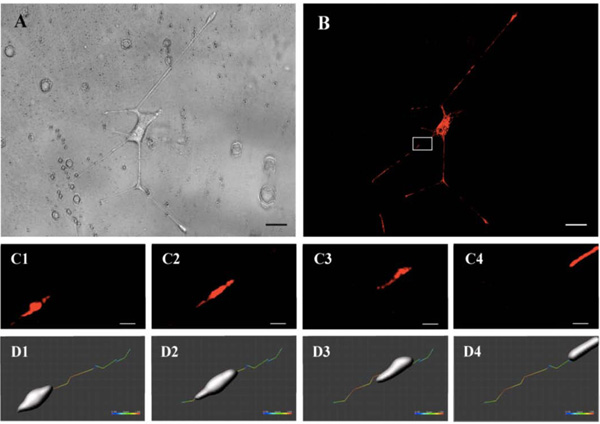
Cell bodies, neurites, and mitochondria were analyzed over several periods of time ranging from minutes to 4-5 hours (movie available on supplementary data: film mitochondria). The acquired images allowed a speed quantification of these structures. Fig. (2A (brightfield) and 2B (fluorescence)) show a PC12 cell body with neurites extended along the surface grating after 5 days. Images were acquired every minute for a period of 30 minutes. Fig. (2C) shows the movement of mitochondria for 15 seconds in the white square of Fig. (2B). Modeling of the organelle at each time point using the module surface of Imaris 8.0 software (Biplane, Zurich, Switzerland) allowed to estimate the trajectory and speed of the mitochondria over the period, ranging from 0.1 to 1 µm/s (Fig. 2D).
3.3. Specific Procedures for the Preservation of Surface Patterns after Preparation for SEM and AFM Observations
We aimed at improving the visualization of cells/bioazopolymer interactions using SEM in order to study the mechanisms involved in the spatial control of cell growth. As cells preparation for SEM microscopy needs exposure to several solvents that can potentially alter or destroy the organic material, including the morphology and structure of the surface photo-induced structuration, we first performed a test to assess the pattern stability under standard dehydration procedure. A surface pattern was photoinduced and scanned via AFM before (Fig. 3A) and after (Fig. 3B) exposure to solvents used in standard fixation procedure. Fig. (3B) shows that the patterned surface of the thin film almost completely disappeared after treatment.
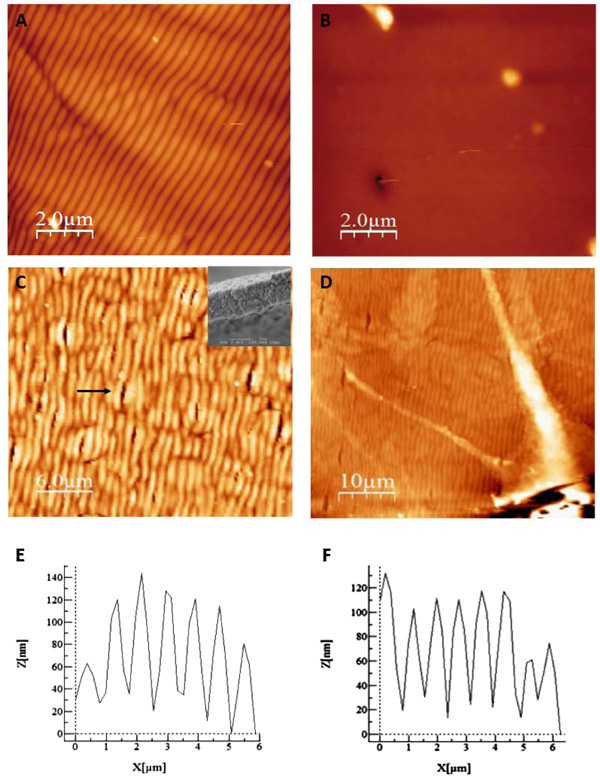
After testing several protocols, we finally ended up by selecting a procedure that preserved the surface relief patterns after treatment (described in Methods). Basically, after conventional preparation, ethanol was replaced by non-diluted propanol for the dehydration. As shown in Fig. (3C), the general organization of the network, assessed using AFM, was preserved after the procedure. The inset in the Fig. (3C) shows that the pitch and amplitude of the surface pattern were also preserved. The surface pattern was intact, only presenting few cracks (arrow on Fig. (3C)) resulting from the local interaction of the products with the surface. These cracks were 500 nm width and variable length and did not perturb the seeding and development of PC12 cells.
We then performed PC12 cells differentiation induced by NGF on our samples. AFM analysis allowed to visualize PC12 cells and their neuritis Fig. (3D). The analysis of the morphological parameters of the grating, depth and width, showed that they were preserved after the procedure using NGF Fig. (3E) before and Fig. (3F) after the procedure. The average pitch and amplitude of the surface pattern modulation before and after the treatment for cell growth was comparable with a difference of 5% regarding the amplitude.
3.4. Analysis of Cells/Azopolymer Interactions using SEM.
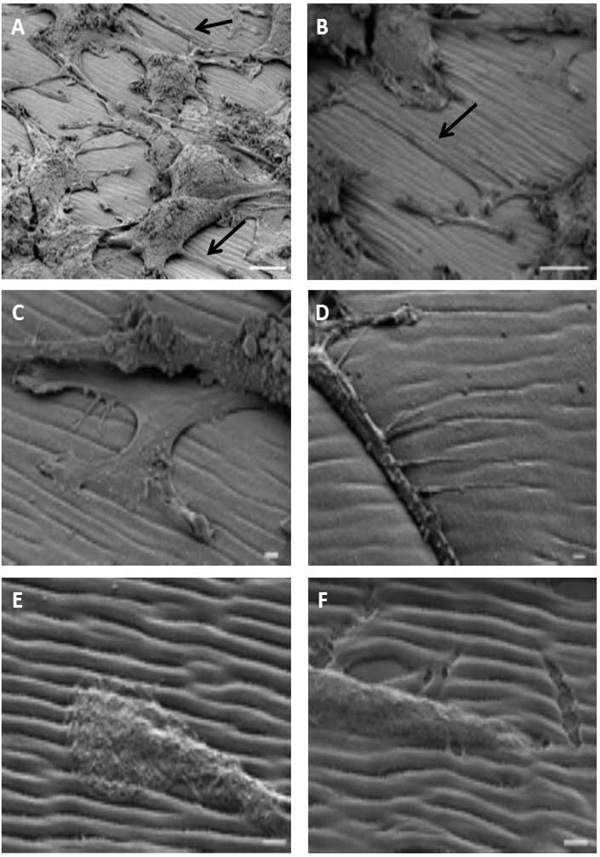
We analyzed PC12 cells seeded on the polymer, differentiated by NGF and prepared with our modified protocol in the vacuum chamber of the microscope, after titling the sample around 45°. As shown on the Fig. (4A), the morphology of the cells was preserved and neurites expanding from the cell bodies along the grating could be observed (arrows on panel A and B, Fig. (4). Neurites followed the groove of the polymer (panel C and D, Fig. (4)). At higher magnification, we observed that the tip of the neurites was guided by the groove (panel E and F on Fig. (4)). Finally, as it was suspected by AFM data, the organization (morphology, depth, width…) of the spontaneous surface relief grating was preserved and allowed a fine study of neurite interactions with the substrate in particular the sensitivity of cells to the mechanical properties of the extracellular matrix.
4. DISCUSSION
Extracellular matrix is a key element influencing the somatotopic development of cells and, so far, of organs. The 3D organization of ECM has high mechanical and biological interactions with cells, which in turn can regulate and reconfigure the ECM [1, 2, 14]. Studies of innermost relationships between cells and ECM are of paramount importance to better appreciate the development, organization and regeneration of organs [5, 7]. In this effort, we developed a reconfigurable azopolymer and adapted several techniques allowing to follow cells from microscopic time-lapse experiments to more precise electronic techniques as SEM. Our results highlight intimate relationships between this 2D artificial ECM and cells extensions, particular the growth cones [2, 14].
2D and 3D artificial ECM are known to influence cell development depending on their structures. Indeed, the same cell line can adopt different comportment if seeded on 2D or 3D matrices [1, 2, 4]. Here we show that PC12 cells respond to the nanometric groove and moreover, instead of having multiple neurites, develop one predominant neurite along the pattern. These results indicate that the biocompatible azopolymer can control the phenotype of neurons as described in our previous results [10]. Furthermore, we were able to conduct time-lapse experiments allowing us to follow the extension of the neurites over time. These results are of importance as they can give insight to the dynamic interactions between ECM and cells without fixing them [2, 6]. This monitoring of living cells can also be very useful to study the dynamic of other cell structures, such as cytoskeleton or vesicles [1, 7, 14]. Our azopolymer thus influences cells growth and architecture and it is suitable for time-lapse experiments, allowing the study of neurites extension through the growth cone [1, 14]. This is a key feature, as most of the time, cells seeded on other usual ECM have to be fixed [2, 15].
Another limit of artificial ECMs, is the impossibility to use electronic techniques to study cells/ECM interactions at higher magnification [2, 4, 15]. In this study, we were able to develop a rapid and efficient protocol that allowed us to use both AFM and SEM [7]. Our SEM results showed, as previously described [4], that neurites follow most of the time the groove of the ECM and that the polymer has no consequences on cells viability and proliferation. Others experiments using different substrates also showed that organization of the ECM helped and enhanced neurites extension [3, 16]. Moreover, the interaction between very small neurites extensions, known as filopodia could easily be seen and observed.
SUMMARY AND CONCLUSION
In conclusion, the use of azopolymer allows the rapid and easily reproducible generation of reconfigurable grooves on the surface of coverslips. Cells seeded on this 2D artificial ECM tend to extend following its architecture. Live cells can be visualized in vivo using optical microscopy. However, fixed cells on photostructurable surfaces prepared with a modified SEM preparation procedure can be visualized by scanning electron microscopy techniques and with a near-field microscope opening the possibility to use this photostructured material for further studies as topological influences on cell growth.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
We are grateful to SCIAM (Service Commun d’Imageries et d’Analyses Microscopiques; SFR4208; UNIV Angers) for their technical assistances in SEM and AFM.
R. Barillé received a grant from “LIA CNRS NAPOLI” (NAnoPhotoswitching of Organic materials with Light) and from University Angers for “Neuropolys” (growth of NEUROns on POLYmer Structures)
SUPPLEMENTARY DATA
Movie (Film mitochondria). A video editing has been made, showing first the phase and fluorescence cell. Then, a mitochondria in an axon has been recorded moving back to the cell body. Finally, the reconstruction showed the tracking of mitochondria movement.


