RESEARCH ARTICLE
Content Based Medical Image Retrieval for Accurate Disease Diagnosis
Mayank R. Kapadia1, *, Chirag N. Paunwala2
Article Information
Identifiers and Pagination:
Year: 2021Volume: 15
Issue: Suppl-2, M12
First Page: 235
Last Page: 248
Publisher ID: TOBEJ-15-235
DOI: 10.2174/1874120702115010235
Article History:
Received Date: 22/1/2021Revision Received Date: 26/8/2021
Acceptance Date: 22/9/2021
Electronic publication date: 31/12/2021
Collection year: 2021

open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution 4.0 International Public License (CC-BY 4.0), a copy of which is available at: https://creativecommons.org/licenses/by/4.0/legalcode. This license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Introduction:
Content Based Image Retrieval (CBIR) system is an innovative technology to retrieve images from various media types. One of the CBIR applications is Content Based Medical Image Retrieval (CBMIR). The image retrieval system retrieves the most similar images from the historical cases, and such systems can only support the physician's decision to diagnose a disease. To extract the useful features from the query image for linking similar types of images is the major challenge in the CBIR domain. The Convolution Neural Network (CNN) can overcome the drawbacks of traditional algorithms, dependent on the low-level feature extraction technique.
Objective:
The objective of the study is to develop a CNN model with a minimum number of convolution layers and to get the maximum possible accuracy for the CBMIR system. The minimum number of convolution layers reduces the number of mathematical operations and the time for the model's training. It also reduces the number of training parameters, like weights and bias. Thus, it reduces the memory requirement for the model storage. This work mainly focused on developing an optimized CNN model for the CBMIR system. Such systems can only support the physicians' decision to diagnose a disease from the images and retrieve the relevant cases to help the doctor decide the precise treatment.
Methods:
The deep learning-based model is proposed in this paper. The experiment is done with several convolution layers and various optimizers to get the maximum accuracy with a minimum number of convolution layers. Thus, the ten-layer CNN model is developed from scratch and used to derive the training and testing images' features and classify the test image. Once the image class is identified, the most relevant images are determined based on the Euclidean distance between the query features and database features of the identified class. Based on this distance, the most relevant images are displayed from the respective class of images. The general dataset CIFAR10, which has 60,000 images of 10 different classes, and the medical dataset IRMA, which has 2508 images of 9 various classes, have been used to analyze the proposed method. The proposed model is also applied for the medical x-ray image dataset of chest disease and compared with the other pre-trained models.
Results:
The accuracy and the average precision rate are the measurement parameters utilized to compare the proposed model with different machine learning techniques. The accuracy of the proposed model for the CIFAR10 dataset is 93.9%, which is better than the state-of-the-art methods. After the success for the general dataset, the model is also tested for the medical dataset. For the x-ray images of the IRMA dataset, it is 86.53%, which is better than the different pre-trained model results. The model is also tested for the other x-ray dataset, which is utilized to identify chest-related disease. The average precision rate for such a dataset is 97.25%. Also, the proposed model fulfills the major challenge of the semantic gap. The semantic gap of the proposed model for the chest disease dataset is 2.75%, and for the IRMA dataset, it is 13.47%. Also, only ten convolution layers are utilized in the proposed model, which is very small in number compared to the other pre-trained models.
Conclusion:
The proposed technique shows remarkable improvement in performance metrics over CNN-based state-of-the-art methods. It also offers a significant improvement in performance metrics over different pre-trained models for the two different medical x-ray image datasets.
1. INTRODUCTION
Content Based Image Retrieval (CBIR) retrieves a set of images from the big bunch of images. The CBIR is the utmost preferable method over Text-Based Image Retrieval (TBIR), owing to laborious tasks like manual annotation of such a big image bunch [1]. CBIR is one of the research's emerging areas due to the wide use of high definition mobile cameras and a wide range of image generation through it. Along with this source of images, the other source, i.e., medical images, also produces many images, such as X-rays, MRI, CT scan, etc., every day, which provides a big challenge for image retrieval. Content Based Medical Image Retrieval (CBMIR) has enormous significance, especially in the clinical decision and research field, due to the availability of the various patients' historical images in the medical clinic. They have crucial data for the diagnosis of the disease. Also, the retrieved similar type of cases can help to diagnose precisely and to decide proper treatment. CBIR's key goal in medical science is to retrieve the images that are semantically like a given query image for successful treatment depending on the seriousness phase of the malady. Hence, the CBMIR can help in the clinical decision support systems, research, and clinical studies in finding relevant information from large repositories [2].
Computer vision is the domain that extracts useful information from the images, which can help to retrieve the images. The CBIR area's major challenge is to reduce the gap between the low-level feature extracted by computer vision techniques and high-level perceptions understood by humans. This gap is known as the semantic gap [2, 3]. The latest neural networks are deep neural networks where the numbers of hidden layers are more between input and output layers. Initially, hidden layers are used to extract the low-level features like color, edge, or shape from the images. These features are later on provided to the higher-level hidden layers to derive the high-level features. Hence, such a machine learning algorithm can derive the advanced features from the images, and therefore it is possible to overcome the semantic gap problem.
The Convolution Neural Network (CNN) is a deep learning technique. In this technique, the features are extracted automatically from the images without the intervention of the human. It is used in this paper to get the advanced features from the images. It is a typical task to identify the number of CNN layers used to extract the images' valuable features in the CNN-based CBIR system. The proposed algorithm contains ten layers of CNN to derive the useful features of the images. The CNN based image retrieval process is shown in Fig. (1). The proposed algorithm enhances retrieval accuracy. It is compared with some other state-of-the-art machine learning methods. The proposed algorithm is also used for the medical image retrieval application, and its results are also compared with some recent machine learning based models.
The proposed work is related to improving the retrieval accuracy by selecting the best tuning parameters of the machine learning techniques for the large image dataset. In addition, the proposed algorithm is also tested for the medical images to show the CBIR's usefulness in medical science.
In the starting stage of CBIR research, some researchers have used low-level features like color features, shape features, and texture features to retrieve the images. However, these features were also used for image retrieval for the different applications of medical science. The various computer vision-based techniques are discussed in Table 1 [4-14].
The CBIR's significant challenges are the semantic gap between these low-level features extracted by computer vision techniques and high-level concepts understood by humans. These computer vision techniques were unable to reduce this semantic gap. In the last few years, many researchers have used machine learning algorithms for image retrieval, which are discussed in Table 2 [15-36].
These machine learning techniques can extract advanced features from images. These advanced features will help to improve the classification accuracy and hence reduce the semantic gap. Some researchers have used CBIR in the field of medical science to diagnose the diseases from the images and retrieve the relevant cases to help the doctor decide the precise treatment. It is also helpful for medical students to learn about the relevant cases and their treatment.
| Technique | Pros. and Cons. |
|---|---|
| Morphology [4], Co-occurrence features [5] | It was used to express the color features but has limitations of the more retrieval time. |
| Color Histogram [6] | For early detection and timely treatment of Diabetic Retinopathy, this technique was developed. It helped to reduce its growth and prevent blindness. The color histogram retrieval system in HSV color space will provide better performance than in RGB color space for Diabetic Retinopathy Detection. However, the accuracy of this method was too limited. |
| Radial Inverse Force Histograms [7] | This technique was useful and effective in retinal disorder’s diagnosis. This technique was developed explicitly for diabetic eye identification. |
| Combination of the color histogram and GLCM [8] Combination of color moments and edge histogram [9] |
This technique was developed to take the benefits of more features to retrieve images. A combination of multiple low-level features helped to improve retrieval accuracy. Still, by the fusion of more features, the retrieval accuracy can be further enhanced. |
| Combination of a histogram and DWT for texture features [10] | Various distance measurement parameters were used to analyze the image retrieval rate for X-ray images. It has been observed that the geometrical and statistical distance measures showed promising results. |
| K-means clustering technique [11-13] | The classification technique was combined with computer vision technology to improve accuracy and retrieval speed. |
| SVM classifier [14] | The SVM classifier's different kernels were used to improve the image retrieval accuracy for the small dataset, and color moments were used to improve the retrieval speed. |
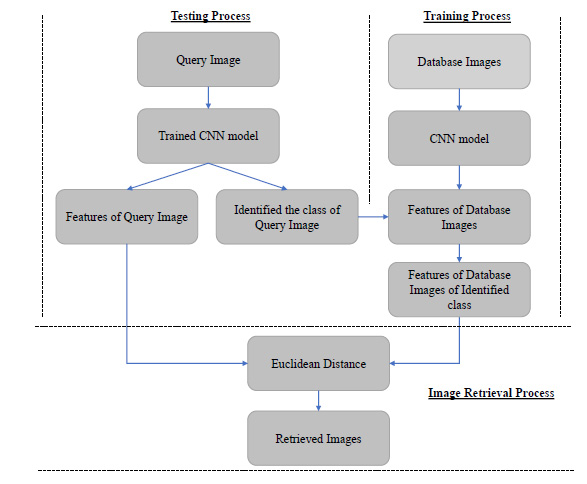 |
Fig (1). CNN based CBIR system. |
| Technique | Pros. and Cons. |
|---|---|
| Root bilinear CNN model [15] | This technique used a square root pooling layer to reduce the feature matrix's size and the memory size and improve the retrieval speed. The performance for Manhattan distance was degraded for the feature size less than 32 |
| Combination of CNN model and PCA model [16] | Feature matrix was reduced by the PCA, which helped to reduce the mathematical complexity, but at the same time, it increased the loss and reduced retrieval accuracy. |
| Pre-trained CNN model [17] | The CNN-based model gives better results than the local descriptor-based methods. The result has been checked for three different general datasets. The CNN fine-tuning strategy can help to improve retrieval efficiency |
| VGG16 model [18, 19] | It provided an excellent retrieval result for only similar images [18] while it could not take advantage of multi-core processing units like GPU [19]. |
| VGG19 model + Closed Form Metric Learning (CFML) [20] | The transfer learning method was used to retrieve the MR images (MRI) for brain tumor retrieval. The medical image dataset is typically small, and training deep CNN for such a small dataset is complicated due to overfitting and convergence problems. |
| CaffeNet model [21] | It was used by adding an extra pooling layer to diminish the number of feature parameters and reduce the computational cost. |
| Transfer learning method [22] | The transfer learning technique was used to retrieve the mammographic images. Hence, it proved that transfer learning could be utilized from a general dataset to a specific medical dataset. |
| CNN model [23, 24] | Used a stochastic pooling layer to overcome the overfitting problem during the training of the large image dataset [23]. The Bayesian optimization method was used to improve the speed of performance [24]. |
| Deep Neural Network (DNN) [25, 26] | Multi-column DNN was used to reduce the semantic gap, improve the classification accuracy for large datasets [25], and reduce the testing error [26]. |
| Micro Neural Network [27] | It helped to extract more abstract features. |
| Hash code + CNN model [28, 29] | This technique used hash like a binary code to retrieve images fast for the large image dataset [28]. In addition, the binary hash code was used to effectively retrieve the retinal images to treat Diabetic Retinopathy (DR) [29]. |
| Generative Adversarial Network [30] | Unsupervised Adversarial Image Retrieval (UAIR) technology was used to retrieve unlabeled images using GAN. The inputs of GAN are noises and realistic images, while CBIR inputs are query images and gallery images. The objective of GAN is to synthesize images that are similar to natural images, while CBIR aims to retrieve well-matched images. Due to the difference between input and output for CBIR and GAN, it is challenging to apply adversarial training directly to CBIR |
| GoogleNet and Additive Latent Semantic Layer (ALSL) [31] | ALSL was used to understand the representation of image features. The efficiency is still needed to investigate for the vast image dataset. |
| Alexnet, GoogleNet, and ResNet50 [32] | They were used to take advantage of the previously trained network to improve accuracy. The complex images often create confusion for the network. It was not easy to detect and recognize such types of images |
| Radon barcode+ Region of Interest (ROI)+ CNN [33] | The method was used for the fast retrieval of the x-ray images. A combination of CNN and Radon barcode was used for fast medical image retrieval, but it needed some benchmark datasets for more validation |
| Radon transform + CNN model [34] | The CNN model was used to extract features from the radiology images, and radon transform was used to shrink the feature map. |
| ROI+ DenseNet model [35] | The spinal x-ray images were used to detect the gender of the person. They had used the CNN model developed from scratch, and the same algorithm was tested by providing ROI images, which improved the retrieval results. Later, the ImageNet pre-trained DensNet model was also used, which achieved much better performance. |
| 3D-CapsNets and a convolutional neural network (CNN) with 3D-autoencoder [36] | The technique was used to detect Alzheimer’s disease from Magnetic resonance imaging (MRI). The CapsNet is made up of a 3D-Convolutional Neural Network and pre-trained 3D-autoencoder technology. As a result, the performance of the hybrid technology was more compared to Deep-CNN alone. Even for a small dataset, CapsNet can learn fast and effectively handle the image robustness. |
| Layer | Type | Input | Output | Parameters |
|---|---|---|---|---|
| Input | Image Input | 3x32x32 | 3x32x32 | 0 |
| Conv2d_1 | Convolution | 3x32x32 | 32x32x32 | 896 |
| ReLU_1 | Activation | 32x32x32 | 32x32x32 | 0 |
| Normalization_1 | Batch normalization | 32x32x32 | 32x32x32 | 128 |
| Conv2d_2 | Convolution | 32x32x32 | 64x32x32 | 18496 |
| ReLU_2 | Activation | 32x32x32 | 64x32x32 | 0 |
| Normalization_2 | Batch normalization | 32x32x32 | 64x32x32 | 256 |
| Pooling2d_1 | Max Pooling | 64x32x32 | 64x16x16 | 0 |
| dropout_1 | Dropout | 64x16x16 | 64x16x16 | 0 |
| Conv2d_3 | Convolution | 64x16x16 | 64x16x16 | 36928 |
| ReLU_3 | Activation | 64x16x16 | 64x16x16 | 0 |
| Normalization_3 | Batch normalization | 64x16x16 | 64x16x16 | 256 |
| Conv2d_4 | Convolution | 64x16x16 | 128x16x16 | 73856 |
| ReLU_4 | Activation | 128x16x16 | 128x16x16 | 0 |
| Normalization_4 | Batch normalization | 128x16x16 | 128x16x16 | 512 |
| Pooling2d_2 | Max Pooling | 128x16x16 | 128x8x8 | 0 |
| dropout_2 | Dropout | 128x8x8 | 128x8x8 | 0 |
| Conv2d_5 | Convolution | 128x8x8 | 128x8x8 | 147584 |
| ReLU_5 | Activation | 128x8x8 | 128x8x8 | 0 |
| Normalization_5 | Batch normalization | 128x8x8 | 128x8x8 | 512 |
| Conv2d_6 | Convolution | 128x8x8 | 256x8x8 | 295168 |
| ReLU_6 | Activation | 256x8x8 | 256x8x8 | 0 |
| Normalization_6 | Batch normalization | 256x8x8 | 256x8x8 | 1024 |
| Pooling2d_3 | Max Pooling | 256x8x8 | 256x4x4 | 0 |
| dropout_3 | Dropout | 256x4x4 | 256x4x4 | 0 |
| Conv2d_7 | Convolution | 256x4x4 | 256x4x4 | 590080 |
| ReLU_7 | Activation | 256x4x4 | 256x4x4 | 0 |
| Normalization_7 | Batch normalization | 256x4x4 | 256x4x4 | 1024 |
| Conv2d_8 | Convolution | 256x4x4 | 512x4x4 | 1180160 |
| ReLU_8 | Activation | 512x4x4 | 512x4x4 | 0 |
| Normalization_8 | Batch normalization | 512x4x4 | 512x4x4 | 2048 |
| Pooling2d_4 | Max Pooling | 512x4x4 | 512x2x2 | 0 |
| dropout_4 | Dropout | 512x2x2 | 512x2x2 | 0 |
| Conv2d_9 | Convolution | 512x2x2 | 512x2x2 | 2359808 |
| ReLU_9 | Activation | 512x2x2 | 512x2x2 | 0 |
| Normalization_9 | Batch normalization | 512x2x2 | 512x2x2 | 2048 |
| Conv2d_10 | Convolution | 512x2x2 | 512x2x2 | 2359808 |
| ReLU_10 | Activation | 512x2x2 | 512x2x2 | 0 |
| Normalization_10 | Batch normalization | 512x2x2 | 512x2x2 | 2048 |
| Pooling2d_5 | Max Pooling | 512x2x2 | 512x1x1 | 0 |
| dropout_5 | Dropout | 512x1x1 | 512x1x1 | 0 |
| Flatten_1 | Flatten | 512x1x1 | 512 | 0 |
| Dense_1 | Dense | 512 | 10 | 5130 |
Further paper is ordered as follows. First, the main architecture of the proposed algorithm is presented in section 2. Then, the quantitative and qualitative study of the proposed algorithm and its comparison with the state-of-the-art method is made in section 3. One of the applications of the CBIR in the domain of medical science is also analyzed in section 3. Finally, the paper is completed with the conclusion.
2. METHODS
2.1. Convolution Neural Network Architecture
The deep learning algorithm uses more hidden layers between input and output compared to the conventional neural network. More hidden layers can learn more abstract features from the input. The deep learning algorithm also requires a vast amount of data for the initial training. In the CBIR's initial technology, the features were extracted and then provided to the classifier for the classification. But the deep learning-based method is an end to end process where high-level features are directly extracted from the images. The deep learning method takes much time for the training data, but once the network is trained, it will take less time for the testing data. The Convolution Neural Network (CNN) is the most popular network of deep learning technology, widely used for 2-D data-like images.
The proposed method for the CBIR is CNN, with many different types of layers. The convolution layer is made up of neurons. They can learn by modifying the weights and biases. While the neural network is being trained, each filter of this convolution layer is convolved with the input image, performs the dot product operation between filter and input image, and generates the 2-dimensional feature map as a result. ReLU is the non-linear activation function that is used after the convolution process. This layer improves the non-linear characteristics of the network. The max-pooling layers are utilized to decrease the spatial dimension of the feature matrix. It is used to decrease the computational cost by directly reducing the number of parameters. This layer also helps to overcome the chances of the overfitting problem of the network. The fully connected layer needs the one-dimensional vector as an input. The last pooling layer's output is given to the flattening layer, which converts the multidimensional matrix into the one-dimensional vector. These layers work as the features extractor, which links the input image's pixel information with its category. The parameters of these layers are tuned and optimized to reduce the misclassification error. The fully connected layer contains a full connection. So, the output of the flattening layer is applied as an input to the fully connected layer. The last layer of the fully connected network performs the classification operation using the softmax activation function because one neuron is available for each class in this layer.
Apart from this, some additional layers like dropout layers are used to overcome the problem of overfitting. In this dropout layer, some neurons are dropped for the training during the training of the network. Hence, mathematical complexity is reduced, and it prevents the overfitting issue of the network. Furthermore, the spatial transformer unit is used to accomplish the geometric transformation of the provided input images. Hence, there is no need to perform data augmentation like translation, scaling, rotation, and skewing manually. Finally, the batch normalization layer is used to normalize the layer's inputs for each mini-batch to stabilize the learning process, which will reduce the number of epoch required to train the network. In this way, batch normalization accelerates the training process and minimizes the generalization error.
Fig. (2) illustrates the architecture of the CNN model. There is an ‘N’ number of convolution layers in between inputs and the flattening layers.
2.2. Proposed Architecture
The CNN architecture is set to perform the classification of the images and measure image retrieval performance. It is shown in Table 3.
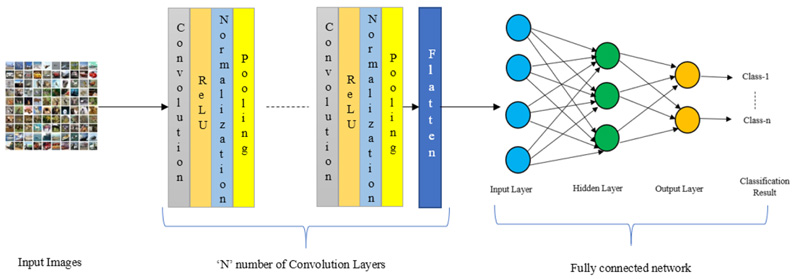 |
Fig. (2). CNN architecture. |
The 10-layer CNN model is being used in the experiment for the classification of the images. The input of the model is an RGB image of 32 x 32 sizes. It will be passed from the different layers, as shown in Table 3, and generate the 512 features at the flatten layer. These features are further used to train the fully connected network for the classification. In the entire architecture, 2x2 max pooling is utilized. The batch normalization is also one of the essential layers in the architecture. It normalizes the input so that the mean output is zero and the standard deviation is one. Thus, it helps to increase the network training speed and also improves the learning rate. The dropout layer is also used to overcome the problem of overfitting. Optimizer is also an essential parameter to reduce the error between the input image's predicted class and actual class. The various experiment is also done to select the best optimizer. The Adam, RMSprop, Nadam, SGD, Adamax, Adagrad, and Adadelata are some examples of optimizers. After the query image classification, the most similar images of the identified class must be retrieved. This process of feature extraction and retrieval of the image is shown in Algorithm 1.
As shown in Algorithm 1, once the query image class has been detected, features of the query image are compared with the database images' features. Based on that, the Euclidean distance between these two is calculated. Finally, the Euclidean distance between the query and dataset images is computed using Eq. 1,
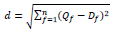 |
(1) |
Here, d is the Euclidean distance between the query image and database images of the identified class. Qf is a feature vector of the query image, and Df is a feature vector of the identified class database image, and f is the number of features. The same class images are organized in ascending order based on this Euclidean distance(d), and top ‘k’ images are retrieved for the provided query image.
2.3. Measurement Parameters
The network has been trained on google collaboratory, which provides free Tesla K80 GPU. Python programming is used in these experiments. The KERAS framework is used for the development of the model. It utilized TensorFlow as the backend. The retrieval accuracy is the term, which is used to evaluate the proposed CNN model.
Accuracy is the ratio of the total number of precise predictions to the total number of predictions. For a single query image, top relevant images are displayed, and the precision rate is calculated to measure the retrieval accuracy. The total number of similar images retrieved to the total number of images retrieved is known as the precision rate. The precision rate is used to calculate the retrieval accuracy of any query image, and it is given by Eq. 2,
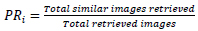 |
(2) |
The average of all the precision rate derived for each query image is known as the average precision rate, and it is given by Eq. 3,
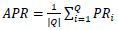 |
(3) |
| Input: Query image, Dataset images | |||
| Output: Retrieved most similar images | |||
| 1 | I ← Query Image | ||
| 2 | Q ← Count of dataset images | ||
| 3 | Initialization of variable: Assign 1 to variable j and k. | ||
| 4 | begin | ||
| 5 | while (j<=Q) do //Read each database image one by one. | ||
| 6 | FQ ←Extract features of the image j at the last flattening layer. | ||
| 7 | end | ||
| 8 | FI ← Extract features of the query image I at the last flattening layer. | ||
| 9 | while (k<=Q) do // calculate the distance | ||
| 10 | d ← Measure Euclidean distance (FI, FQ) | ||
| 11 | end | ||
| 12 | Top ranked images ← sort(d) | ||
| 13 | end | ||
PRi is the ith query image's precision rate, and Q is the total number of the query image. In our experiment, once the image class is correctly identified, then the identical images are retrieved from the same image class. So, the average precision rate gives the same result as classification accuracy.
3. RESULTS AND DISCUSSION
3.1. CBIR for the Non-Medical Dataset
The proposed CNN model is mainly designed for the medical image datasets. Still, to check the accuracy and the perfection of the model, it is also tested for the well-known and widely used non-medical ‘CIFAR10’ dataset [37]. ‘CIFAR10’ dataset is widely used for most CBIR systems, and CBMIR is one of the CBIR applications. Therefore, a non-medical ‘CIFAR10’ dataset is tested to check the versatility of the proposed model. The ‘CIFAR-10’ dataset consists of 60,000 images of ten different classes, where 6000 images are available in each category. Based on the object, the classes are formed. The objects are airplane, automobile, bird, cat, deer, dog, frog, horse, ship, and truck. The images are the RGB images whose size is 32 x32. Images of the different classes have been shown in Fig. (3).
The images of all the classes are distributed into training, validation, and testing sets. Based on this division, 40,000 images are used for the model's training, 10,000 are used as the validation set, and 10,000 are used as the testing set. All the images are different in the training and testing dataset. For the whole bunch of the images, the JPEG format is being used.
Initially, to develop the CNN model from scratch, the layers are varied to identify the minimum number of layers required for the deep network for the best accuracy. Table 4 shows the simulation result by applying different CNN layers.
 |
Fig. (3). Sample images of each class. |
| Accuracy | No. of CNN Layers = 6 | No. of CNN Layers= 8 | No. of CNN Layers= 10 | No. of CNN Layers= 11 |
|---|---|---|---|---|
| Training | 87.72% | 87.74% | 95.03% | 94.89% |
| Validation | 86.55% | 87.94% | 90.21% | 90.16% |
| Testing | 87.72% | 87.96% | 89.90% | 89.59% |
After many trials, it has been observed that there is no significant increase in the accuracy even if the layers are increasing. So, as the number of layers increases, it increases the time for training. Here, 10-layers give good training and testing accuracy. Hence, ten number of CNN layers are adopted for the designing of the CNN model. After deciding the number of layers for the CNN, the following experiment selects another fine-tuning parameter, batch size. Table 5 shows the simulation result by applying different batch sizes.
| Accuracy | Batch Size | |||
|---|---|---|---|---|
| 100 | 200 | 300 | 400 | |
| Training | 92.12% | 93.60% | 92.61% | 95.03% |
| Validation | 89.34% | 89.74% | 89.78% | 90.21% |
| Testing | 88.41% | 89.43% | 88.93% | 89.90% |
Here, as shown in Table 5, the accuracy is maximum for 400 batch size. In addition, it has been observed that when batch size increases, the time required to train the network decreases.
Next, the experiment is performed to select the CNN model's best optimizer. The optimizer is one of the hyperparameters of the CNN model. It is responsible for reducing the error between the actual class and the predicted class of the training images. Tuning the trainable parameters like weights and bias improves the training accuracy and minimizes the training loss. For selecting the best optimizer for the proposed CNN model, the experiment is done with different optimizers by keeping the same values for the number of convolution layers, epochs, and batch size.
Fig. (4) shows the analysis of the classification accuracy of the proposed model for the different optimizers. Here, the values of the accuracy are mentioned in the attached table in the same figure. It shows that the accuracy of Adadelta, Adagrad, and SGD is not up to the mark. However, it also shows that the accuracy of Adam, RMSprop, Nadam, and Adamax is almost nearer to each other. The RMSprop has the highest validation and testing accuracy of 89.76% and 89.38%, respectively. Adamax has the highest training accuracy of 98.19%.
Fig. (5) shows the training time required for the different optimizers. The testing accuracy of the Adam, RMSprop, Nadam, and Adamax is almost nearer to each other, as shown in Fig. (4), but as per Fig. (5), the training time required using RMSprop is 12% to 22% less than the other optimizers. Also, the RMSprop has the highest validation and testing accuracy of 89.76% and 89.38%, respectively. So, from these experiments, the CNN model with RMSprop optimizer provides a good result.
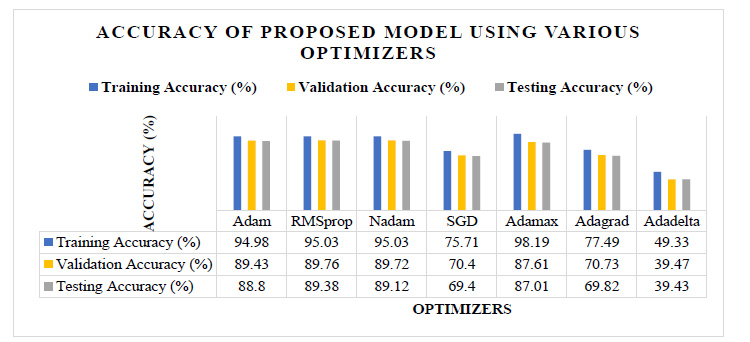 |
Fig. (4). Accuracy of the proposed model using various optimizers. |
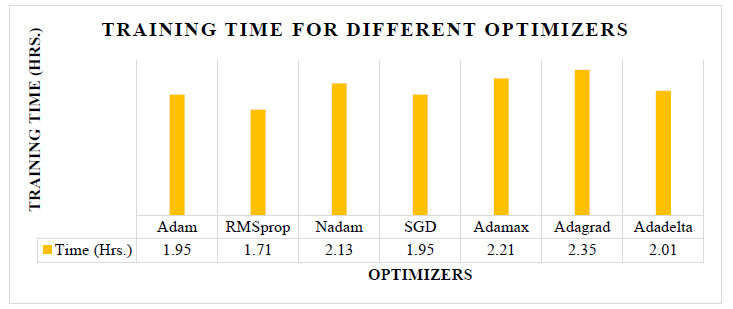 |
Fig. (5). Analysis of training time for different Optimizers. |
| Name of Hyperparameters | Value of Hyperparameter |
|---|---|
| Number of Convolution Layers | 10 |
| Batch Size | 400 |
| Optimizer | RMSprop |
| Epochs | 300 |
The proposed architecture listed in Table 3 is trained with 400 batch size and RMSprop optimizers. Fig. (6) shows the accuracy curve and the loss curve for the proposed CNN model. Fig. (6a) shows that the training and validation accuracy increases with the increase in epochs, while as shown in Fig. (6b), the training and validation loss decreases with the increase in epochs. After 300 epochs, it has been observed that the accuracy and loss become constant, and no further increase of epochs is required.
The optimization by LDWPSO can find excellent hyperparameters and can provide high classification accuracy [38]. For example, the accuracy of the proposed model with the mentioned parameters in Table 6 is 93.9%, far better than the 69.37% accuracy of the LDWPSO CNN model [38] for the CIFAR-10 dataset.
The proposed CNN model’s results are compared with the different CNN-based models implemented in the literature [23, 25, 27, 30]. These are the methods where tuning parameters were modified. Fig. (7) shows the comparative analysis of the proposed model's classification accuracy over the CIFAR10 dataset with the other CNN-based methods implemented in the literature.
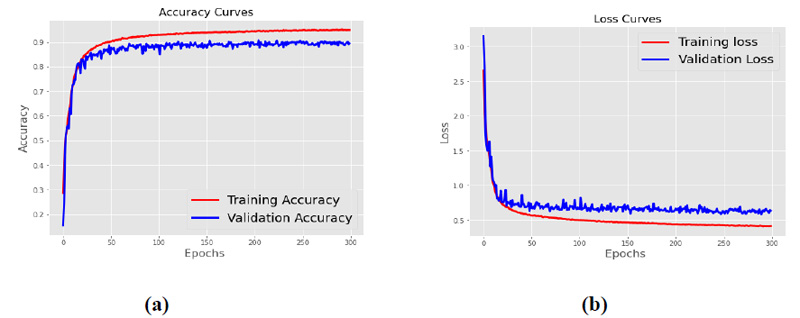 |
Fig. (6). (a) Accuracy curve (b) Loss curve. |
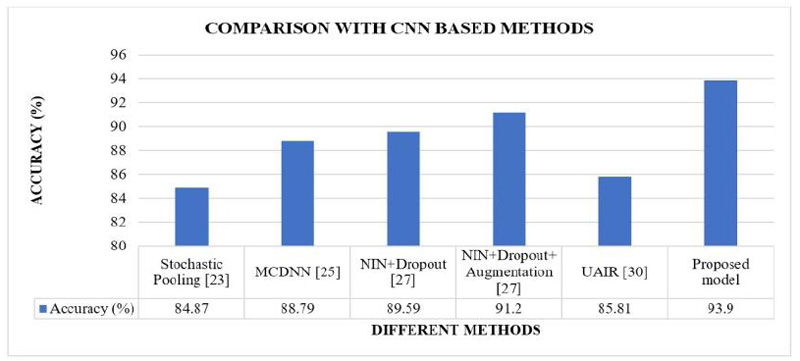 |
Fig. (7). Comparison of different CNN based methods. |
The proposed CNN model’s results are also compared with the different CNN-based hybrid models implemented in the literature [24, 28, 31]. These are the methods where CNN models were combined with some other techniques. Fig. (8) shows the comparative analysis of the proposed model's classification accuracy over the CIFAR10 dataset with the other CNN-based hybrid methods implemented in the literature.
The proposed CNN model’s results are also compared with the different transfer learning methods implemented in the literature [32]. These are the methods where existing CNN models were fine-tuned. Fig. (10) shows the comparative analysis of the proposed model's classification accuracy over the CIFAR10 dataset with the other transfer learning-based methods implemented in the literature.
Figs. (7 and 8) show that our proposed model's accuracy is 93.9%, better than the different CNN-based models and different CNN-based hybrid models. Fig (9) shows that our proposed model's accuracy is far better than some transfer learning methods like Alexnet, GoogleNet, and ResNet50. The above results are summarized in Table 7.
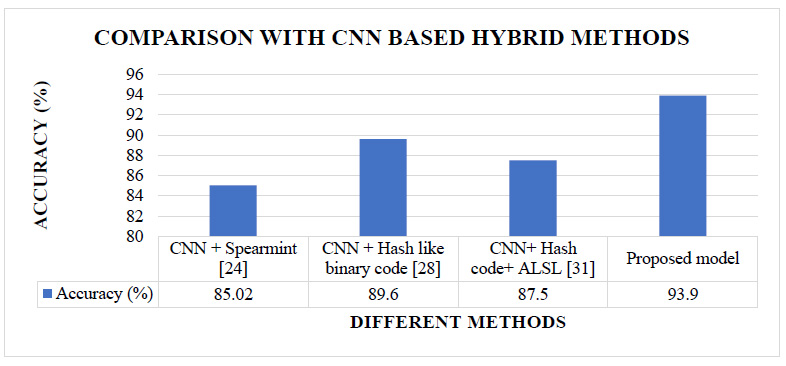 |
Fig. (8). Comparison with different CNN based Hybrid methods. |
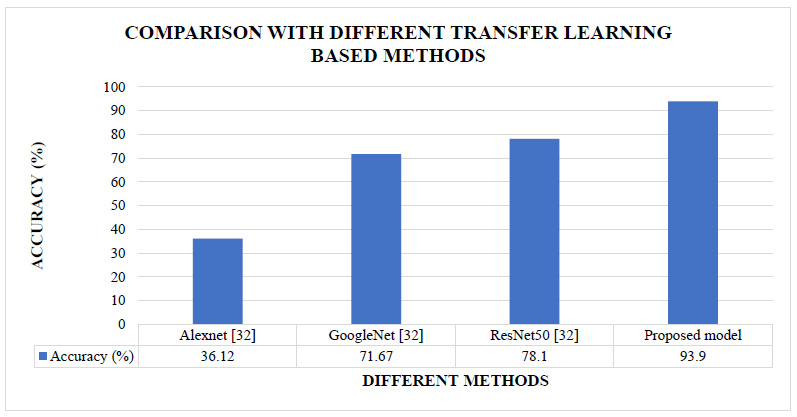 |
Fig. (9). Comparison with the different transfer learning methods. |
 |
Fig. (10). (a) Query Image (b) Retrieval resultant images. |
| Type of Methods | Methods | %Improvement in Accuracy Over Existing Methods |
|---|---|---|
| CNN based method | Stochastic Pooling [23] | 11% |
| MCDNN [25] | 6% | |
| NIN+Dropout [27] | 5% | |
| NIN+Dropout+Augmentation [27] | 3% | |
| UAIR [30] | 9% | |
| Hybrid method | CNN + Spearmint [24] | 10% |
| CNN + Hash like binary code [28] | 5% | |
| CNN+ Hash code+ ALSL [31] | 7% | |
| Transfer learning method | Alexnet [32] | 160% |
| GoogleNet [32] | 31% | |
| ResNet50 [32] | 20% |
Table 7 shows that the proposed model improves the accuracy from 3% to 160% over the other existing methods. CNN provides statistically better performance than the conventional hand-crafted feature extraction techniques [38]. Also, a smaller number of convolution layers are required to extract the high-level features for the small dataset to reduce the semantic gap [39]. Similarly, a few more convolution layers are needed for a vast dataset to get the high-level features to overcome the semantic gap problem. The relationship between the semantic gap and classification accuracy can be given by Eq. 4.
 |
(4) |
So, from Eq. 4, the semantic gap can be minimized by increasing the classification accuracy. Hence, the semantic gap for the proposed model for the non-medical CIFAR10 dataset is 6.1%.
Here, versatility of the proposed model is tested for the benchmark non-medical “CIFAR10” dataset. Its performance is good compared to other popular techniques described in Figs. (7-9). Some state-of-art methods [40] achieve acceptable accuracy for the CIFAR10 dataset, but the number of convolution layers utilized in those techniques is higher. Hence, it required more training time and needed more memory to store the trainable parameters. The proposed model gives maximum accuracy with a minimum number of convolution layers and minimum trainable parameters.
Once the query image class is recognized, then similar images can be retrieved from the database by calculating the Euclidean distance between the query image features and the database images' features. The most similar images are retrieved based on the value of the Euclidean distance. For example, Fig. (10) shows the retrieval result for the query image from the ‘car’ class.
As shown in Fig. (10a), a car's image is provided as a query image to the CBIR system. As shown in Fig. (10b), the system has retrieved the top 6 images based on the Euclidean distance between the feature maps of the query image and the feature maps of the database images.
3.2. CBIR for Medical Dataset.
One of the applications of CBIR is Content Based Medical Image Retrieval (CBMIR). The proposed algorithm is also tested for the application of medical image retrieval. The proposed model is developed from scratch, and the proposed model is not using any ImageNet pre-trained models. Therefore, it can learn equally from non-medical and medical datasets. For the CBMIR task, two different datasets have been used. The dataset that contains X-ray images of the chest section of the human body regarding various chest-related diseases has been obtained from Dr. Milind Parekh, Homeopathic Practitioner, Bardoli. The dataset contains 1090 images of 512x512 size. Based on the disease related to the chest, the dataset is divided into five different classes. The various diseases are Tuberculosis, COVID, Normal chest, Pleural effusion, and Viral Pneumonia. All these diseases can be diagnosed using the chest's x-ray image and used for the CBMIR experiment. Images of the different classes have been shown in Fig. (11).
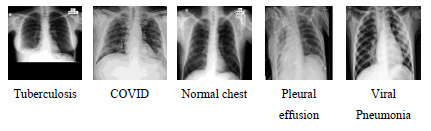 |
Fig. (11). Sample images of the chest dataset for different disease. |
The proposed algorithm is also tested for another medical image dataset. To assess the performance of the proposed model, 2508 x-ray images of the different classes have been used from the standard dataset called Image Retrieval Medical Applications (IRMA) dataset. After sending an agreement signed by the concerned authority, the IRMA database has been received from Dr. Thomas Deserno, Department of Medical Informatics, Aachen University of Technology, Germany. Based on the body parts, the dataset is divided into nine different classes. The different classes' names are Abdomen, Chest, Head, Hip, Leg, Mouth, Neck, Spine, and Upper extremity. Images of the different classes have been shown in Fig. (12).
In this experiment, all the images are distributed into training, validation, and testing sets. Sixty percent of the images are used as training images, 20% are used as validation images, and the remaining 20% are used as testing images. Once the model is trained, it will be tested with the non-trained testing dataset, compared with the actual ground truth to validate the proposed CNN model's decision. The transfer learning technique is used to train the pre-trained model. Transfer learning is a method to reuse the trained model for different applications. Transfer learning is used to save training time with good performance. Here, the network's early convolutional layers are frozen to train only the last few layers, e.g., fully connected layers, making a prediction. Here, during the training, the last three layers are trained for all the pre-trained models. The pre-trained models are the benchmark models for image classification. Hence, the proposed architecture results are compared with the various ImageNet pre-trained models for the medical image datasets, as shown in Table 8.
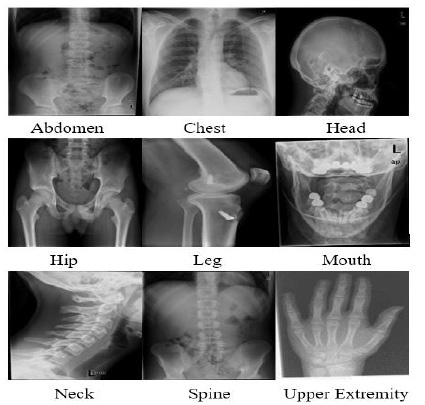 |
Fig. (12). Sample images of the IRMA dataset for different class. |
| Model Name | Number of CNN layers in the model | Average Precision Rate (APR) (%) for Chest Dataset | Average Precision Rate (APR) (%) for IRMA x-ray dataset |
|---|---|---|---|
| AlexNet | 8 | 20.18 | 20.8 |
| VGG16 | 16 | 93.12 | 84.6 |
| VGG19 | 19 | 94.04 | 82.2 |
| ResNet50 | 50 | 92.2 | 82.2 |
| ResNet101 | 101 | 89.45 | 82.2 |
| ResNet152 | 152 | 90.37 | 81.2 |
| ResNet50V2 | 50 | 33.03 | 36.6 |
| ResNet101V2 | 101 | 32.57 | 64.6 |
| ResNet152V2 | 152 | 29.36 | 50.2 |
| MobileNet | 28 | 69.27 | 70.4 |
| MobileNetV2 | 28 | 60.09 | 65.2 |
| DenseNet121 | 121 | 93.12 | 83.4 |
| Dense Net169 | 169 | 93.12 | 82 |
| DenseNet201 | 201 | 77.98 | 82.2 |
| Proposed Model | 10 | 97.25 | 86.53 |
Table 8 shows that the proposed model's Average Precision Rate (APR) is 97.25% for the chest disease dataset and 86.53% for the IRMA dataset, which is better than the different ImageNet pre-trained models. The semantic gap of the proposed model for the chest disease dataset is 2.75%, and for the IRMA dataset, it is 13.47%. As shown in Table 5, the proposed model presents the least value of the semantic gap compared to the other popular methods. Only the AlexNet model used fewer CNN layers than the proposed model, but the APR of AlexNet is too much less than the proposed model for both the medical image dataset. The rest of the pre-trained model used many CNN layers compared to the proposed model. But for both the medical image dataset, the proposed model's average precision rate is far better than them. The proposed model is optimized in terms of the number of convolution layers required to get good accuracy.
Here, as shown in Fig. (13a) image of a ‘COVID’ disease from the chest dataset is provided as a query image to the CBMIR system, and as shown in Fig. (13b), the system has retrieved the top 16 images related to the ‘COVID’ disease. In addition, the system also retrieved the historical cases and their related treatment of the disease.
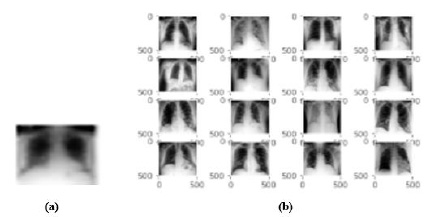 |
Fig. (13). (a) Query Image of ‘COVID’ disease (b) Retrieval resultant images for ‘COVID’ disease. |
Fig. (14a) shows the X-ray image of the ‘spine’ provided as a query image to the CBMIR system, and the system retrieves the top 16 images related to the spine class, as shown in Fig. (14b). The CBMIR system's output does not retrieve images of any other body parts that qualitatively validate the classification performance. The system also retrieved the historical cases and their details related to the images.
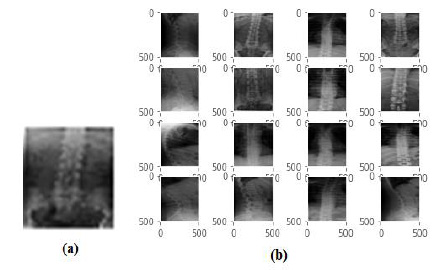 |
Fig. (14). (a) Query Image of Spine class, (b) Retrieval resultant images for Spine class. |
CONCLUSION
CNN can extract a high level of features, and due to that, it can be used to reduce the semantic gap between the low-level features and high-level concepts. The experimental result also shows that the proposed method's accuracy in retrieving the images is better than the literature’s method. The batch normalization layer of the CNN model is used to extract more useful features and stabilize the learning process, decreasing the number of epochs required to train the network. Dropout is also used in the network to temporarily deactivate some neurons' learning to improve the learning speed and reduce the over fitting problem. The more significant number of layers are utilized to derive the more abstract features to retrieve the image precisely. The batch size is also playing an essential role in the improvement in retrieval accuracy. The RMSprop optimizer provides good accuracy and takes less time for training than different available optimizers. The vast dataset requires many convolutional layers to derive the more abstract features for good retrieval performance. The results also conclude that the maximum accuracy has been achieved with the smaller layers than some other pre-trained models. This good result motivates the researchers to apply this algorithm to develop some real-time applications like Content Based Medical Image Retrieval, which can be a supportive tool for clinical diagnosis. The proposed algorithm significantly improves compared to the other different transfer learning models for the two different medical x-ray image datasets. It is observed that the proposed model is giving good performance with a medical image dataset and a non-medical dataset. But it is not being claimed that the proposed model is generalized for all kinds of CBIR applications. This methodology might create CBIR for other body organ MRI pictures and other clinical imaging spaces like PET and CT.
LIST OF ABBREVIATIONS
| CBIR | = Content Based Image Retrieval |
| CBMIR | = Content Based Medical Image Retrieval |
| CNN | = Convolution Neural Network |
| GPU | = Graphics Processing Unit |
| MRI | = Magnetic Resonance Imaging |
| PR | = Precision Rate |
| APR | = Average Precision Rate |
| IRMA | = Image Retrieval Medical Applications |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The authors confirm that the data supporting the findings of this research are available within the article.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Thomas Deserno, Department of Medical Informatics, Aachen University of Technology, Germany, for giving the dataset to do this research work. The authors also might want to offer their heartiest thanks to Dr. Milind Parekh, Hem Homeopathy, Bardoli for his advice and support from a clinical point of view, all through this exploration.
REFERENCES
| [1] | H El-kaime, M Hanoune, and A. Eddaoui, Applying data analytics and Cumulative Accuracy Profile (CAP) Approach, vol. 756, Springer International Publishing, 2019. |
| [2] | A. Qayyum, S.M. Anwar, M. Awais, and M. Majid, "Medical image retrieval using deep convolutional neural network", Neurocomputing, vol. 266, pp. 8-20, 2017. |
| [3] | Q. Zhang, and E. Izquierdo, "Histology image retrieval in optimised multi-feature spaces", IEEE J. Biomed. Health Inform., vol. 17, no. 1, pp. 240-249, 2013. |
| [4] | E. Aptoula, and S. Lefèvre, "Morphological description of color images for content-based image retrieval", IEEE Trans. Image Process., vol. 18, no. 11, pp. 2505-2517, 2009. |
| [5] | A.P. Bhagat, and M. Atique, "Design and development of systems for image segmentation and content based image retrieval", 2nd Natl Conf Comput Intell Signal Process CISP 2012, pp. 109-113, 2012. |
| [6] | N. Kashyap, and D.K. Singh, "Color histogram based image retrieval technique for diabetic retinopathy detection", 2nd Int Conf Converg Technol I2CT 2017, pp. 799-802, 2017. |
| [7] | S. Kimpan, N. Maneerat, and C. Kimpan, "Diabetic retinopathy image analysis using radial inverse force histograms. ICIIBMS2017", 2nd Int Conf Intell Informatics Biomed Sci, pp. 266-271, 2018. |
| [8] | J. Yue, Z. Li, L. Liu, and Z. Fu, "Content-based image retrieval using color and texture fused features", Math. Comput. Model., vol. 54, no. 3-4, pp. 1121-1127, 2011. |
| [9] | E. Walia, S. Vesal, and A. Pal, "An effective and fast hybrid framework for color image retrieval", Sens. Imaging, vol. 15, no. 1, 2014. |
| [10] | P. Nalini, and B.L. Malleswari, "Performance of medical image retrieval techniques: A comparative analysis", 4th Int Conf Electron Commun Syst ICECS 2017, pp. 28-32, 2017. |
| [11] | Y.C. Chen, C.S. Sastry, V.M. Patel, P.J. Phillips, and R. Chellappa, "In-plane rotation and scale invariant clustering using dictionaries", IEEE Trans. Image Process., vol. 22, no. 6, pp. 2166-2180, 2013. |
| [12] | R.I. Chang, S.Y. Lin, J.M. Ho, C.W. Fann, and Y.C. Wang, "A novel content based image retrieval system using K-means/KNN with feature extraction", Comput. Sci. Inf. Syst., vol. 9, no. 4, pp. 1645-1661, 2012. |
| [13] | J.F. Serrano-Talamantes, C. Avilés-Cruz, J. Villegas-Cortez, and J.H. Sossa-Azuela, "Self organizing natural scene image retrieval", Expert Syst. Appl., vol. 40, no. 7, pp. 2398-2409, 2013. |
| [14] | M.R. Kapadia, and C.N. Paunwala, "Analysis of SVM kernels for content based image retrieval system", Int Conf Energy, Commun Data Anal Soft Comput ICECDS 2017, pp. 1409-1414, 2017. |
| [15] | A. Alzu’bi, A. Amira, and N. Ramzan, "Content-based image retrieval with compact deep convolutional features", Neurocomputing, vol. 249, pp. 95-105, 2017. |
| [16] | L. Wang, and X. Wang, "Model and metric choice of image retrieval system based on deep learning", Proc - 2016 9th Int Congr Image Signal Process Biomed Eng Informatics, CISP-BMEI 2016, pp. 390-395, 2016. |
| [17] | L. Zheng, Y. Yang, and Q. Tian, "SIFT meets CNN: A decade survey of instance retrieval", IEEE Trans. Pattern Anal. Mach. Intell., vol. 40, no. 5, pp. 1224-1244, 2018. |
| [18] | Z. Rian, V. Christanti, and J. Hendryli, "Content-based image retrieval using Convolutional Neural Networks", Proc - 2019 IEEE Int Conf Signals Syst ICSigSys 2019, pp. 1-7, 2019. |
| [19] | F. Liu, Y. Wang, F-C. Wang, Y-Z. Zhang, and J. Lin, "Intelligent and secure content-based image retrieval for mobile users", IEEE Access, vol. 7, pp. 119209-119222, 2019. |
| [20] | Z.N.K. Swati, Q. Zhao, and M. Kabir, "Content-based brain tumor retrieval for MR images using transfer learning", IEEE Access, vol. 7, no. c, pp. 17809-17822, 2019. |
| [21] | M Tzelepi, and A. Tefas, "IEEE 13th image, Video, Multidimens Signal Process Work IVMSP", Proc, vol. 1-5, 2018. |
| [22] | R.S. Bressan, D.H.A. Alves, L.M. Valerio, P.H. Bugatti, and P.T.M. Saito, "Doctor: The role of deep features in content-based mammographic image retrieval", Proc - IEEE Symp Comput Med Syst., pp. 158-163, 2018. |
| [23] | MD Zeiler, and R Fergus, "Stochastic pooling for regularization of deep convolutional neural networks arXiv", |
| [24] | J. Snoek, H. Larochelle, and R.P. Adams, "Practical bayesian optimization of machine learning algorithms", Adv. Neural Inf. Process. Syst., pp. 2951-2959, 2012. |
| [25] | D Cires, and U. Meier, Multi-column deep neural networks for image classification., 2012, pp. 3642-3649. |
| [26] | A. Krizhevsky, and G.E. Hinton, "ImageNet classification with deep convolutional neural networks", Adv. Neural Inf. Process. Syst., pp. 1097-1105, 2012. |
| [27] | M. Lin, Q. Chen, and S Yan, "Network in network", arXiv., . Available from: https://arxiv.org/abs/1312.4400 |
| [28] | K Lin, H Yang, J Hsiao, and C. Chen, Deep learning of binary hash codes for fast image retrieval., 2015, pp. 27-35. |
| [29] | I. Wijesinghe, C. Gamage, and C. Chitraranjan, "Deep supervised hashing through ensemble cnn feature extraction and low-rank matrix factorization for retinal image retrieval of diabetic retinopathy", Proc - 2019 IEEE 19th Int Conf Bioinforma Bioeng BIBE, pp. 301-308, 2019. |
| [30] | S. Chen, and Y. Lu, "Adversarial learning for content-based image retrieval", IEEE Conf Multimed Inf Process Retr, pp. 97-102, 2019. |
| [31] | J Li, and J. Li, Prompt image search with deep convolutional neural network via efficient hashing code and addictive latent semantic layer., vol. 19, 2018, no. 3, pp. 949-958. |
| [32] | N Sharma, and N. Sharma, "An neural analysis of convolutional neural networks for image classification", Procedia Comput Sci, vol. 132, no. Iccids), pp. 377-384, 2018. |
| [33] | X Liu, HR Tizhoosh, and J Kofman, "Generating binary tags for fast medical image retrieval on convolutional nets and radon transform", 2016 International Joint Conference on Neural Networks (IJCNN), 2016, pp. 2872-2878. |
| [34] | A Khatami, M Babaie, A Khosravi, HR Tizhoosh, SM Salaken, and S Nahavandi, "A deep-structural medical image classification for a radon-based image retrieval", Can Conf Electr Comput Eng., pp. 17-20, 2017. |
| [35] | Z. Xue, S. Rajaraman, R. Long, S. Antani, and G. Thoma, "Gender detection from spine x-ray images using deep learning", Proc - IEEE Symp Comput Med Syst, pp. 54-58, 2018. |
| [36] | KR Kruthika, Rajeswari, and HD. Maheshappa, "Erratum: CBIR system using Capsule Networks and 3D CNN for Alzheimer’s disease diagnosis", Informatics Med Unlocked, vol. 14, pp. 59-68, 2019. |
| [37] | A Krizhevsky, and GE Hinton, Learning multiple layers of features from tiny images., 2009. |
| [38] | T. Serizawa, and H Fujita, "Optimization of convolutional neural network using the linearly decreasing weight particle swarm optimization", arXiv. Available from: http://arxiv.org/abs/2001.05670 |
| [39] | M.R. Kapadia, and C.N. Paunwala, "Improved CBIR system using Multilayer CNN", Int. Conf. Inventive Research in Computing App., ICIRCA, pp. 840-845, 2018. |
| [40] | Image Classification on CIFAR-10, available from: https://paperswithcode.com/sota/image-classification-on-cifar-10 |








